Temporal Ecological Niche Models.
tenm 
An R package with a set of functions to calibrate time-specific ecological niche models. Time-specific niche modeling (TENM) is a novel approach that allows calibrating niche models with high temporal resolution spatial information, which aims to reduce niche estimation biases. Although TENM could improve distribution estimates, few works have used them. The goal of tenm R package is to provide methods and functions to calibrate time-specific niche models, letting users execute a strict calibration and selection process of niche models based on ellipsoids, as well as functions to project the potential distribution in the present and in global change scenarios.
Installation
You can install the development version of tenm from GitHub with:
if (!require('devtools')) install.packages('devtools')
devtools::install_github("luismurao/tenm")
# If you want to build vignette, install pandoc before and then
devtools::install_github('luismurao/tenm',build_vignettes=TRUE)
Example
We start with a simple example to show the basic functions of the package. We will work with a dataset of Abronia graminea, an endemic lizard from the Mexican Sierra Madre Oriental.
First, we load the tenm R package.
library(tenm)
## basic example code
Now we load the abronia dataset, which contains geographical information about the presence of Abronia graminea in its area of distribution. This dataset has also information about the year of observation and the GBIF doi.
data("abronia")
head(abronia)
#> species decimalLongitude decimalLatitude year
#> 1 Abronia graminea -98.17773 19.96523 2014
#> 2 Abronia graminea -98.13753 19.87006 2014
#> 3 Abronia graminea -98.07042 19.89668 2014
#> 4 Abronia graminea -98.13003 19.86861 2014
#> 5 Abronia graminea -98.14894 19.84450 2014
#> 6 Abronia graminea -98.15909 19.86878 2014
#> gbif_doi
#> 1 https://doi.org/10.15468/dl.teyjm9
#> 2 https://doi.org/10.15468/dl.teyjm9
#> 3 https://doi.org/10.15468/dl.teyjm9
#> 4 https://doi.org/10.15468/dl.teyjm9
#> 5 https://doi.org/10.15468/dl.teyjm9
#> 6 https://doi.org/10.15468/dl.teyjm9
dim(abronia)
#> [1] 106 5
We plot the geographic information to see how Abronia graminea is distributed.
colorss <- hcl.colors(length(unique(abronia$year)))
par(mar=c(4,4,2,2))
plot(abronia$decimalLongitude, abronia$decimalLatitude,
col=colorss,pch=19, cex=0.75,
xlab="Longitude",ylab="Latitude",xlim=c(-98.35,-96.7))
legend("bottomleft",legend = sort(unique(abronia$year))[1:20],
cex=0.85,pt.cex = 1,bty = "n",
pch=19,col =colorss[1:20])
legend("bottomright",
legend = sort(unique(abronia$year))[21:length(unique(abronia$year))],
cex=0.85,pt.cex = 1,bty = "n",
pch=19,col =colorss[21:length(unique(abronia$year))])
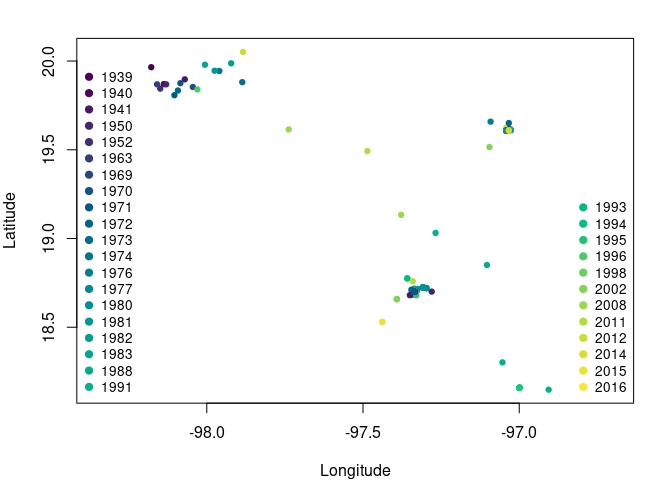
Fig. 1. Occurrence points of *Abronia graminea*. Colors represent the year of observation.
Note that some occurrences are overlapped but belong to different years.
Standard data thinning
A relevant step when curating occurrence data is to eliminate duplicated geographical information, which depends on several factors, including spatial autocorrelation and the spatial resolution of the modeling layers. Let’s see what happens when we eliminate duplicated information as defined by the spatial resolution of our modeling layers. To do this, we will use the tenm::clean_dup function of the tenm R package.
# Load a modeling layer
tempora_layers_dir <- system.file("extdata/bio",package = "tenm")
tenm_mask <- terra::rast(file.path(tempora_layers_dir,"1939/bio_01.tif"))
ab_1 <- tenm::clean_dup(data =abronia,
longitude = "decimalLongitude",
latitude = "decimalLatitude",
threshold = terra::res(tenm_mask),
by_mask = FALSE,
raster_mask = NULL)
tidyr::as_tibble(ab_1)
#> # A tibble: 10 × 5
#> species decimalLongitude decimalLatitude year gbif_doi
#> <chr> <dbl> <dbl> <int> <chr>
#> 1 Abronia graminea -97.5 19.5 1995 https://doi.org/10.1…
#> 2 Abronia graminea -97.0 18.2 1993 https://doi.org/10.1…
#> 3 Abronia graminea -98.0 19.8 1980 https://doi.org/10.1…
#> 4 Abronia graminea -97.7 19.6 2012 https://doi.org/10.1…
#> 5 Abronia graminea -97.9 20.1 2015 https://doi.org/10.1…
#> 6 Abronia graminea -97.4 18.5 1952 https://doi.org/10.1…
#> 7 Abronia graminea -97.1 18.9 1998 https://doi.org/10.1…
#> 8 Abronia graminea -97.3 19.0 1983 https://doi.org/10.1…
#> 9 Abronia graminea -97.3 18.7 1973 https://doi.org/10.1…
#> 10 Abronia graminea -97.0 19.7 1972 https://doi.org/10.1…
After applying our spatial thinning, we obtained only ten observations from 106 occurrences. We lost a lot of information!!! The function tenm::clean_dup has a method to clean duplicated records according to a raster mask layer. The above avoids losing records that might occur in different pixels but fall within the distance used as threshold for cleaning.
ab_by_mask <- tenm::clean_dup(data =abronia,
longitude = "decimalLongitude",
latitude = "decimalLatitude",
threshold = terra::res(tenm_mask),
by_mask = TRUE,
raster_mask = tenm_mask)
tidyr::as_tibble(ab_by_mask)
#> # A tibble: 16 × 5
#> species decimalLongitude decimalLatitude year gbif_doi
#> <chr> <dbl> <dbl> <int> <chr>
#> 1 Abronia graminea -98.2 20.0 2014 https://doi.org/10.1…
#> 2 Abronia graminea -98.1 19.9 2014 https://doi.org/10.1…
#> 3 Abronia graminea -98.1 19.8 2014 https://doi.org/10.1…
#> 4 Abronia graminea -97.9 19.9 2014 https://doi.org/10.1…
#> 5 Abronia graminea -97.3 18.7 1963 https://doi.org/10.1…
#> 6 Abronia graminea -97.1 18.3 1996 https://doi.org/10.1…
#> 7 Abronia graminea -97.4 18.8 1941 https://doi.org/10.1…
#> 8 Abronia graminea -97.4 18.7 1988 https://doi.org/10.1…
#> 9 Abronia graminea -97.0 19.6 1991 https://doi.org/10.1…
#> 10 Abronia graminea -97.4 19.1 2002 https://doi.org/10.1…
#> 11 Abronia graminea -97.5 19.5 1995 https://doi.org/10.1…
#> 12 Abronia graminea -97.0 18.2 1993 https://doi.org/10.1…
#> 13 Abronia graminea -97.7 19.6 2012 https://doi.org/10.1…
#> 14 Abronia graminea -97.9 20.1 2015 https://doi.org/10.1…
#> 15 Abronia graminea -97.1 18.9 1998 https://doi.org/10.1…
#> 16 Abronia graminea -97.3 19.0 1983 https://doi.org/10.1…
We recovered 6 records, not bad! On the other hand, we did not account for the fact that some occurrences come from different years. The tenm package is designed to deal with occurrences coming from different periods as long as the user has environmental layers matching the years of occurrence observations.
Time-specific niche modeling
Let’s apply the functions and methods to work with time-specific niche models. First, we load our data.
library(tenm)
data("abronia")
Now, we indicate the path where our time-specific modeling layers are located.
tempora_layers_dir <- system.file("extdata/bio",package = "tenm")
print(tempora_layers_dir)
#> [1] "/home/luis/R/x86_64-pc-linux-gnu-library/4.4/tenm/extdata/bio"
We explore the structure of the directory that contains our modeling layers.
list.dirs(tempora_layers_dir,recursive = FALSE)
#> [1] "/home/luis/R/x86_64-pc-linux-gnu-library/4.4/tenm/extdata/bio/1939"
#> [2] "/home/luis/R/x86_64-pc-linux-gnu-library/4.4/tenm/extdata/bio/1940"
#> [3] "/home/luis/R/x86_64-pc-linux-gnu-library/4.4/tenm/extdata/bio/1941"
#> [4] "/home/luis/R/x86_64-pc-linux-gnu-library/4.4/tenm/extdata/bio/1950"
#> [5] "/home/luis/R/x86_64-pc-linux-gnu-library/4.4/tenm/extdata/bio/1952"
#> [6] "/home/luis/R/x86_64-pc-linux-gnu-library/4.4/tenm/extdata/bio/1963"
#> [7] "/home/luis/R/x86_64-pc-linux-gnu-library/4.4/tenm/extdata/bio/1969"
#> [8] "/home/luis/R/x86_64-pc-linux-gnu-library/4.4/tenm/extdata/bio/1970"
#> [9] "/home/luis/R/x86_64-pc-linux-gnu-library/4.4/tenm/extdata/bio/1971"
#> [10] "/home/luis/R/x86_64-pc-linux-gnu-library/4.4/tenm/extdata/bio/1972"
#> [11] "/home/luis/R/x86_64-pc-linux-gnu-library/4.4/tenm/extdata/bio/1973"
#> [12] "/home/luis/R/x86_64-pc-linux-gnu-library/4.4/tenm/extdata/bio/1974"
#> [13] "/home/luis/R/x86_64-pc-linux-gnu-library/4.4/tenm/extdata/bio/1976"
#> [14] "/home/luis/R/x86_64-pc-linux-gnu-library/4.4/tenm/extdata/bio/1977"
#> [15] "/home/luis/R/x86_64-pc-linux-gnu-library/4.4/tenm/extdata/bio/1980"
#> [16] "/home/luis/R/x86_64-pc-linux-gnu-library/4.4/tenm/extdata/bio/1981"
#> [17] "/home/luis/R/x86_64-pc-linux-gnu-library/4.4/tenm/extdata/bio/1982"
#> [18] "/home/luis/R/x86_64-pc-linux-gnu-library/4.4/tenm/extdata/bio/1983"
#> [19] "/home/luis/R/x86_64-pc-linux-gnu-library/4.4/tenm/extdata/bio/1988"
#> [20] "/home/luis/R/x86_64-pc-linux-gnu-library/4.4/tenm/extdata/bio/1991"
#> [21] "/home/luis/R/x86_64-pc-linux-gnu-library/4.4/tenm/extdata/bio/1993"
#> [22] "/home/luis/R/x86_64-pc-linux-gnu-library/4.4/tenm/extdata/bio/1994"
#> [23] "/home/luis/R/x86_64-pc-linux-gnu-library/4.4/tenm/extdata/bio/1995"
#> [24] "/home/luis/R/x86_64-pc-linux-gnu-library/4.4/tenm/extdata/bio/1996"
#> [25] "/home/luis/R/x86_64-pc-linux-gnu-library/4.4/tenm/extdata/bio/1998"
#> [26] "/home/luis/R/x86_64-pc-linux-gnu-library/4.4/tenm/extdata/bio/2002"
#> [27] "/home/luis/R/x86_64-pc-linux-gnu-library/4.4/tenm/extdata/bio/2008"
#> [28] "/home/luis/R/x86_64-pc-linux-gnu-library/4.4/tenm/extdata/bio/2011"
#> [29] "/home/luis/R/x86_64-pc-linux-gnu-library/4.4/tenm/extdata/bio/2012"
#> [30] "/home/luis/R/x86_64-pc-linux-gnu-library/4.4/tenm/extdata/bio/2014"
#> [31] "/home/luis/R/x86_64-pc-linux-gnu-library/4.4/tenm/extdata/bio/2015"
#> [32] "/home/luis/R/x86_64-pc-linux-gnu-library/4.4/tenm/extdata/bio/2016"
Note that the directory contains other directories named with the dates of the modeling layers. Now, we explore some of these dated directories.
# Directory for year 1939
list.files(list.dirs(tempora_layers_dir,
recursive = FALSE)[1],
pattern = ".tif$")
#> [1] "bio_01.tif" "bio_02.tif" "bio_03.tif" "bio_04.tif" "bio_05.tif"
#> [6] "bio_06.tif" "bio_07.tif" "bio_08.tif" "bio_09.tif" "bio_10.tif"
#> [11] "bio_11.tif" "bio_12.tif" "bio_13.tif" "bio_14.tif" "bio_15.tif"
#> [16] "bio_16.tif" "bio_17.tif" "bio_18.tif" "bio_19.tif"
# Directory for year 1972
list.files(list.dirs(tempora_layers_dir,
recursive = FALSE)[10],
pattern = ".tif$")
#> [1] "bio_01.tif" "bio_02.tif" "bio_03.tif" "bio_04.tif" "bio_05.tif"
#> [6] "bio_06.tif" "bio_07.tif" "bio_08.tif" "bio_09.tif" "bio_10.tif"
#> [11] "bio_11.tif" "bio_12.tif" "bio_13.tif" "bio_14.tif" "bio_15.tif"
#> [16] "bio_16.tif" "bio_17.tif" "bio_18.tif" "bio_19.tif"
# Directory for year 2014
list.files(list.dirs(tempora_layers_dir,
recursive = FALSE)[30],
pattern = ".tif$")
#> [1] "bio_01.tif" "bio_02.tif" "bio_03.tif" "bio_04.tif" "bio_05.tif"
#> [6] "bio_06.tif" "bio_07.tif" "bio_08.tif" "bio_09.tif" "bio_10.tif"
#> [11] "bio_11.tif" "bio_12.tif" "bio_13.tif" "bio_14.tif" "bio_15.tif"
#> [16] "bio_16.tif" "bio_17.tif" "bio_18.tif" "bio_19.tif"
Note that all dated directories must have the same environmentalinformation. In this example, we used the bioclimatic layers derived from the CHELSAcruts database.
The sp.temporal.modeling object
In the following lines of code, we will use a special function of the tenm R package that will allow us to work with time-specific data.
data("abronia")
tempora_layers_dir <- system.file("extdata/bio",package = "tenm")
abt <- tenm::sp_temporal_data(occs = abronia,
longitude = "decimalLongitude",
latitude = "decimalLatitude",
sp_date_var = "year",
occ_date_format="y",
layers_date_format= "y",
layers_by_date_dir = tempora_layers_dir,
layers_ext="*.tif$")
The function tenm::sp_temporal_data is parameterized with the occurrence dated database. To parameterize the function, we need to specify the name of the columns that contain the longitude and latitude data, the column that represents the year of observation, the format of dates (here years, but see the help of the function for other date formats), the layers date format, the directory that contains the time-specific modeling layers and the raster layer extension.
The object abt is a special class called sp.temporal.modeling that deals with time-specific information.
In the following line of code, we explore the slots of abt object.
# See the names of the slots
names(abt)
#> [1] "temporal_df" "sp_date_var" "lon_lat_vars" "layers_ext"
The abt object has four slots: temporal data.frame (“temporal_df”), a character vector indicating the date variable (“sp_date_var”), a character vector showing the names of longitude and latitude data (“lon_lat_vars”) and another character vector with the extension of the modeling layers.
Now, we explore the temporal_df slot, which is a data.frame with five columns: longitude, latitude, the time variable (here year), the layer dates, and layers path (the path the temporal niche layers are located).
# See the temporal data.frame
tidyr::as_tibble(head(abt$temporal_df))
#> # A tibble: 6 × 5
#> decimalLongitude decimalLatitude year layer_dates layers_path
#> <dbl> <dbl> <int> <date> <chr>
#> 1 -98.2 20.0 2014 2014-01-01 /home/luis/R/x86_64-pc-lin…
#> 2 -98.1 19.9 2014 2014-01-01 /home/luis/R/x86_64-pc-lin…
#> 3 -98.1 19.9 2014 2014-01-01 /home/luis/R/x86_64-pc-lin…
#> 4 -98.1 19.9 2014 2014-01-01 /home/luis/R/x86_64-pc-lin…
#> 5 -98.1 19.8 2014 2014-01-01 /home/luis/R/x86_64-pc-lin…
#> 6 -98.2 19.9 2014 2014-01-01 /home/luis/R/x86_64-pc-lin…
Time-specific spatial data thinning
As a first step, we will curate our time-specific database using the function tenm::clean_dup_by_date. This function is parametrized as the tenm::clean_dup function with the difference that it thins the data considering the time variable (some occurrences might be spatially duplicated but belong to other dates, so in a time-specific context, they are not duplicates).
# Clean duplicates using a raster mask
abtc <- tenm::clean_dup_by_date(this_species = abt,
by_mask = TRUE,
threshold = terra::res(tenm_mask)[1],
raster_mask = tenm_mask[1],
n_ngbs = 0)
# Check number of records
head(tidyr::as_tibble(abtc$temporal_df))
#> # A tibble: 6 × 5
#> decimalLongitude decimalLatitude year layer_dates layers_path
#> <dbl> <dbl> <int> <date> <chr>
#> 1 -97.3 18.7 1939 1939-01-01 /home/luis/R/x86_64-pc-lin…
#> 2 -97.3 18.7 1940 1940-01-01 /home/luis/R/x86_64-pc-lin…
#> 3 -97.0 19.6 1941 1941-01-01 /home/luis/R/x86_64-pc-lin…
#> 4 -97.3 18.7 1941 1941-01-01 /home/luis/R/x86_64-pc-lin…
#> 5 -97.3 18.7 1950 1950-01-01 /home/luis/R/x86_64-pc-lin…
#> 6 -97.1 19.7 1950 1950-01-01 /home/luis/R/x86_64-pc-lin…
nrow(abtc$temporal_df)
#> [1] 40
An improvement of this methodology is that we recover a lot of information. From 10 records thinned using the standard data cleaning process, now we have 40 records; 30 more observations!!! which will allow us to fit more informative models. Let’s compare occurrences from the standard data cleaning process and the time-specific thinning process.
colors1 <- hcl.colors(length(unique(ab_1$year)))
par(mar=c(4,4,2,2),mfrow=c(1,2))
plot(ab_1$decimalLongitude, ab_1$decimalLatitude,
col=colors1,pch=19, cex=0.75,
xlab="Longitude",ylab="Latitude",xlim=c(-98.35,-96.7))
legend("bottomleft",legend = sort(unique(ab_1$year))[1:10],
cex=0.85,pt.cex = 1,bty = "n",
pch=19,col =colors1[1:10])
colors2 <- hcl.colors(length(unique(abtc$temporal_df$year)))
plot(abtc$temporal_df$decimalLongitude, abtc$temporal_df$decimalLatitude,
col=colors2,pch=19, cex=0.75,
xlab="Longitude",ylab="Latitude",xlim=c(-98.35,-96.7))
legend("bottomleft",legend = sort(unique(abtc$temporal_df$year))[1:16],
cex=0.85,pt.cex = 1,bty = "n",
pch=19,col =colors2[1:16])
legend("bottomright",
legend = sort(unique(abronia$year))[17:length(unique(abtc$temporal_df$year))],
cex=0.85,pt.cex = 1,bty = "n",
pch=19,col =colors2[17:length(unique(abtc$temporal_df$year))])
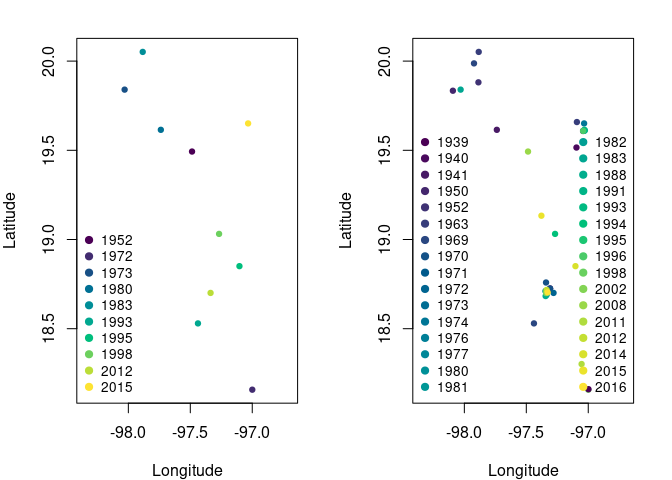
Fig. 2. Comparison of the spatial distribution of occurrence records for the standard thinning processs and the time-specific thinning process. Left panel shows the records after the standard thinning process. Right panel shows the spatial distribution of the records after the time-specific thinning process; note that some records overlap but are from different years.
Time-specific environmental data extraction
After the spatial thinning process, we need to extract environmental information from our occurrence points. The tenm package does this using the function tenm::ex_by_date. This function can be run in parallel by evoking functions of the future package. To parametrize the function, we need to specify the “sp.temporal.modeling” object (obtained using the function tenm::sp_temporal_data or the one from tenm::clean_dup_by_date) and the proportion of occurrences to be used as the training dataset. The tenm package uses a random partition to divide the database into train and test datasets.
future::plan("multisession",workers=2)
abex <- tenm::ex_by_date(this_species = abtc,
train_prop=0.7)
future::plan("sequential")
Now, we explore the slot “temporal_df”.
head(abex$temporal_df)
#> # A tibble: 6 × 26
#> decimalLongitude decimalLatitude year layer_dates layers_path cell_ids_year
#> <dbl> <dbl> <int> <date> <chr> <dbl>
#> 1 -97.3 18.7 1939 1939-01-01 /home/luis/R… 272
#> 2 -97.3 18.7 1940 1940-01-01 /home/luis/R… 272
#> 3 -97.0 19.6 1941 1941-01-01 /home/luis/R… 173
#> 4 -97.3 18.7 1941 1941-01-01 /home/luis/R… 271
#> 5 -97.3 18.7 1950 1950-01-01 /home/luis/R… 272
#> 6 -97.1 19.7 1950 1950-01-01 /home/luis/R… 173
#> # ℹ 20 more variables: bio_01 <int>, bio_02 <int>, bio_03 <int>, bio_04 <int>,
#> # bio_05 <int>, bio_06 <int>, bio_07 <int>, bio_08 <int>, bio_09 <int>,
#> # bio_10 <int>, bio_11 <int>, bio_12 <int>, bio_13 <int>, bio_14 <int>,
#> # bio_15 <int>, bio_16 <int>, bio_17 <int>, bio_18 <int>, bio_19 <int>,
#> # trian_test <chr>
It has 40 rows and columns with the time-specific environmental values and an additional column indicating if the observation will be used as train or test.
Time-specific background generation
The tenm package uses environmental background to compute the ROC and partial ROC test and estimate the prevalence of the species in the environmental space (proportion of environmental points inside the niche model). We will generate 10,000 environmental background points using as calibration area and a neighborhood of 10 pixels around each occurrence point (buffer_ngbs parameter).
future::plan("multisession",workers=2)
abbg <- tenm::bg_by_date(this_species = abex,
buffer_ngbs=10,n_bg=10000)
future::plan("sequential")
head(abbg$env_bg)
#> ID_YEAR
#> 1 /home/luis/R/x86_64-pc-linux-gnu-library/4.4/tenm/extdata/bio/1939
#> 2 /home/luis/R/x86_64-pc-linux-gnu-library/4.4/tenm/extdata/bio/1939
#> 3 /home/luis/R/x86_64-pc-linux-gnu-library/4.4/tenm/extdata/bio/1939
#> 4 /home/luis/R/x86_64-pc-linux-gnu-library/4.4/tenm/extdata/bio/1939
#> 5 /home/luis/R/x86_64-pc-linux-gnu-library/4.4/tenm/extdata/bio/1939
#> 6 /home/luis/R/x86_64-pc-linux-gnu-library/4.4/tenm/extdata/bio/1939
#> decimalLongitude decimalLatitude bio_01 bio_02 bio_03 bio_04 bio_05 bio_06
#> 1 -97.75000 18.91667 155 92 57 2177 223 62
#> 2 -98.25000 18.75000 192 100 60 1990 264 97
#> 3 -98.58333 17.75000 210 104 62 1574 286 118
#> 4 -98.41667 19.41667 134 99 59 2204 205 38
#> 5 -96.58333 17.91667 204 80 53 2221 271 121
#> 6 -98.75000 18.41667 234 105 61 1801 311 139
#> bio_07 bio_08 bio_09 bio_10 bio_11 bio_12 bio_13 bio_14 bio_15 bio_16 bio_17
#> 1 161 175 136 176 124 550 133 1 1 279 9
#> 2 167 210 177 211 163 690 187 0 1 408 7
#> 3 168 223 192 224 187 616 177 1 1 375 5
#> 4 167 156 103 156 103 613 119 0 1 326 8
#> 5 151 224 199 225 173 2520 619 4 1 1167 39
#> 6 172 248 214 250 208 611 158 0 1 380 3
#> bio_18 bio_19
#> 1 259 14
#> 2 338 8
#> 3 296 5
#> 4 277 8
#> 5 972 583
#> 6 269 4
The number of background points for each year is proportional to the number of occurrences for each year of observation.
Exporting time-specific information as Samples With Data format
Although the package uses minimum volume ellipsoids to model the niche, it has a function to export the time-specific data to Samples With Data format table that allows users to fit other algorithms such as MaxEnt. Let’s see how it works.
# SWD table for occurrence records
occ_swd <- tdf2swd(this_species=abex,sp_name="abro_gram")
# SWD table for background data
bg_swd <- tdf2swd(this_species=abbg)
head(tidyr::as_tibble(occ_swd))
#> # A tibble: 6 × 23
#> sp_name decimalLongitude decimalLatitude year bio_01 bio_02 bio_03 bio_04
#> <chr> <dbl> <dbl> <int> <int> <int> <int> <int>
#> 1 abro_gram -97.3 18.7 1939 149 84 55 2252
#> 2 abro_gram -97.3 18.7 1940 154 87 49 2575
#> 3 abro_gram -97.0 19.6 1941 114 63 44 2693
#> 4 abro_gram -97.3 18.7 1941 139 82 50 2394
#> 5 abro_gram -97.3 18.7 1950 155 94 54 2079
#> 6 abro_gram -97.1 19.7 1950 115 79 51 2278
#> # ℹ 15 more variables: bio_05 <int>, bio_06 <int>, bio_07 <int>, bio_08 <int>,
#> # bio_09 <int>, bio_10 <int>, bio_11 <int>, bio_12 <int>, bio_13 <int>,
#> # bio_14 <int>, bio_15 <int>, bio_16 <int>, bio_17 <int>, bio_18 <int>,
#> # bio_19 <int>
head(tidyr::as_tibble(bg_swd))
#> # A tibble: 6 × 23
#> sp_name decimalLongitude decimalLatitude year bio_01 bio_02 bio_03 bio_04
#> <chr> <dbl> <dbl> <dbl> <int> <int> <int> <int>
#> 1 background -97.7 18.9 1939 155 92 57 2177
#> 2 background -98.2 18.8 1939 192 100 60 1990
#> 3 background -98.6 17.8 1939 210 104 62 1574
#> 4 background -98.4 19.4 1939 134 99 59 2204
#> 5 background -96.6 17.9 1939 204 80 53 2221
#> 6 background -98.7 18.4 1939 234 105 61 1801
#> # ℹ 15 more variables: bio_05 <int>, bio_06 <int>, bio_07 <int>, bio_08 <int>,
#> # bio_09 <int>, bio_10 <int>, bio_11 <int>, bio_12 <int>, bio_13 <int>,
#> # bio_14 <int>, bio_15 <int>, bio_16 <int>, bio_17 <int>, bio_18 <int>,
#> # bio_19 <int>
Time-specific model calibration and selection
As a final step, we will calibrate time-specific niche models using minimum volume ellipsoids. To achieve this, we first select the environmental variables using the function tenm::correlation_finder. This function filters variables according to a correlation threshold, which is important to avoid issues related to collinearity.
varcorrs <- tenm::correlation_finder(environmental_data =
abex$env_data[,-ncol(abex$env_data)],
method = "spearman",
threshold = 0.8,
verbose = FALSE)
#> Warning in stats::cor(environmental_data, method = method): La desviación
#> estándar es cero
# Selected variables
vars2fit <- varcorrs$descriptors
print(vars2fit)
#> [1] "bio_01" "bio_02" "bio_03" "bio_04" "bio_07" "bio_12" "bio_14" "bio_15"
#> [9] "bio_17"
Now, we use the function tenm::tenm_selection to calibrate the time-specific niche models. This function uses the background object (here, the abbg object) as input. To parametrize the function, we need to specify the omission rate criteria to be used to select the models, the proportion of points to be included in the ellipsoid model (ellipsoid_level parameter), the names of the modeling layers (vars2fit parameter), a numeric vector indicating the number of dimensions used to build ellipsoid models (vars2fit parameter) a logical argument that determines whether to use the partial ROC test or not, the random percent of data to be used for the bootstrap of the partial ROC test (RandomPercent parameter), the number of iterations of the partial ROC test (NoOfIteration parameter), a logical argument to specify whether to run the calibration process in parallel and the number of cores used in the parallel process (parallel parameter).
mod_sel <- tenm::tenm_selection(this_species = abbg,
omr_criteria =0.1,
ellipsoid_level=0.975,
vars2fit = vars2fit,
nvars_to_fit=c(2,3,4,5,6,7),
proc = T,
RandomPercent = 50,
NoOfIteration=1000,
parallel=TRUE,
n_cores=4)
#> -------------------------------------------------------------------
#> **** Starting model selection process ****
#> -------------------------------------------------------------------
#>
#> A total number of 36 models will be created for combinations of 9 variables taken by 2
#>
#> A total number of 84 models will be created for combinations of 9 variables taken by 3
#>
#> A total number of 126 models will be created for combinations of 9 variables taken by 4
#>
#> A total number of 126 models will be created for combinations of 9 variables taken by 5
#>
#> A total number of 84 models will be created for combinations of 9 variables taken by 6
#>
#> A total number of 36 models will be created for combinations of 9 variables taken by 7
#>
#> -------------------------------------------------------------------
#> **A total number of 492 models will be tested **
#>
#> -------------------------------------------------------------------
#> Doing calibration from model 1 to 100 in process 1
#>
#> Doing calibration from model 101 to 200 in process 2
#>
#> Doing calibration from model 201 to 300 in process 3
#>
#> Doing calibration from model 301 to 400 in process 4
#>
#> Doing calibration from model 401 to 492 in process 5
#>
#> Finishing calibration of models 1 to 100
#>
#> Finishing calibration of models 101 to 200
#>
#> Finishing calibration of models 201 to 300
#>
#> Finishing calibration of models 301 to 400
#>
#> Finishing calibration of models 401 to 492
#>
#> Finishing...
#>
#> -------------------------------------------------------------------
#> 244 models passed omr_criteria for train data
#> 27 models passed omr_criteria for test data
#> 27 models passed omr_criteria for train and test data
We fitted 492 models, from which 27 passed our selection criteria. Let’s explore the mod_sel object.
names(mod_sel)
#> [1] "temporal_df" "sp_date_var" "lon_lat_vars" "layers_ext" "env_bg"
#> [6] "mods_table"
It has five slots. We can obtain the table of results of the selection process by calling the mods_table slot.
head(mod_sel$mods_table,27)
#> fitted_vars nvars om_rate_train non_pred_train_ids
#> 1 bio_01,bio_02,bio_04,bio_07 4 0.06250 18,31
#> 2 bio_01,bio_02,bio_03,bio_04 4 0.06250 18,31
#> 3 bio_01,bio_03,bio_04,bio_07 4 0.06250 18,31
#> 4 bio_01,bio_04,bio_07,bio_12 4 0.09375 21,28,31
#> 5 bio_01,bio_02,bio_03,bio_07 4 0.03125 18
#> 6 bio_01,bio_04,bio_07 3 0.06250 18,31
#> 7 bio_01,bio_03,bio_04,bio_12 4 0.09375 18,21,28
#> 8 bio_01,bio_03,bio_04 3 0.06250 3,18
#> 9 bio_01,bio_04 2 0.03125 18
#> 10 bio_01,bio_02,bio_04 3 0.09375 3,18,31
#> 11 bio_01,bio_07 2 0.06250 18,31
#> 12 bio_01,bio_03 2 0.06250 3,18
#> 13 bio_01,bio_02 2 0.09375 3,18,31
#> 14 bio_01,bio_03,bio_12 3 0.06250 3,18
#> 15 bio_02,bio_04,bio_07,bio_12 4 0.06250 21,28
#> 16 bio_02,bio_03,bio_07,bio_12 4 0.06250 21,28
#> 17 bio_02,bio_03,bio_04,bio_12 4 0.06250 21,28
#> 18 bio_01,bio_07,bio_12 3 0.06250 18,31
#> 19 bio_02,bio_03,bio_04,bio_07 4 0.03125 3
#> 20 bio_04,bio_07 2 0.03125 3
#> 21 bio_04,bio_07,bio_12 3 0.06250 21,28
#> 22 bio_04,bio_12 2 0.06250 10,21
#> 23 bio_03,bio_04 2 0.03125 3
#> 24 bio_02,bio_07,bio_12 3 0.09375 3,21,28
#> 25 bio_02,bio_03,bio_12 3 0.09375 3,21,28
#> 26 bio_03,bio_07,bio_12 3 0.09375 3,21,28
#> 27 bio_07,bio_12 2 0.03125 28
#> om_rate_test non_pred_test_ids bg_prevalence pval_bin pval_proc
#> 1 0 0.4706024 0 0
#> 2 0 0.4554527 0 0
#> 3 0 0.4639894 0 0
#> 4 0 0.4627871 0 0
#> 5 0 0.4088013 0 0
#> 6 0 0.4648311 0 0
#> 7 0 0.4625466 0 0
#> 8 0 0.4655525 0 0
#> 9 0 0.4835878 0 0
#> 10 0 0.4768546 0 0
#> 11 0 0.5001804 0 0
#> 12 0 0.4995792 0 0
#> 13 0 0.4823855 0 0
#> 14 0 0.4844295 0 0
#> 15 0 0.6570879 0 0
#> 16 0 0.5706385 0 0
#> 17 0 0.6412168 0 0
#> 18 0 0.4962126 0 0
#> 19 0 0.6100757 0 0
#> 20 0 0.7537574 0 0
#> 21 0 0.6847421 0 0
#> 22 0 0.7220151 0 0
#> 23 0 0.7382470 0 0
#> 24 0 0.6783696 0 0
#> 25 0 0.6816160 0 0
#> 26 0 0.7000120 0 0
#> 27 0 0.7766021 0 0
#> env_bg_paucratio env_bg_auc mean_omr_train_test rank_by_omr_train_test
#> 1 1.520879 0.7985825 0.031250 11
#> 2 1.505433 0.7996963 0.031250 7
#> 3 1.500235 0.7974812 0.031250 8
#> 4 1.467522 0.7828762 0.046875 22
#> 5 1.465208 0.7519788 0.015625 1
#> 6 1.462961 0.7858312 0.031250 9
#> 7 1.448599 0.7549675 0.046875 21
#> 8 1.431383 0.7479400 0.031250 10
#> 9 1.429927 0.7371300 0.015625 2
#> 10 1.424194 0.7454525 0.046875 23
#> 11 1.409348 0.7345675 0.031250 15
#> 12 1.409145 0.7019625 0.031250 14
#> 13 1.398036 0.7285350 0.046875 24
#> 14 1.382147 0.6807450 0.031250 12
#> 15 1.379969 0.7115000 0.031250 18
#> 16 1.374939 0.6537675 0.031250 16
#> 17 1.373252 0.7036713 0.031250 17
#> 18 1.365017 0.7032725 0.031250 13
#> 19 1.329613 0.6943437 0.015625 3
#> 20 1.307749 0.6769513 0.015625 5
#> 21 1.303573 0.6712913 0.031250 19
#> 22 1.288655 0.6388300 0.031250 20
#> 23 1.267623 0.6472100 0.015625 4
#> 24 1.261588 0.6146950 0.046875 25
#> 25 1.253496 0.5970800 0.046875 26
#> 26 1.240056 0.5891425 0.046875 27
#> 27 1.193064 0.5648688 0.015625 6
#> rank_omr_aucratio
#> 1 1
#> 2 2
#> 3 3
#> 4 4
#> 5 5
#> 6 6
#> 7 7
#> 8 8
#> 9 9
#> 10 10
#> 11 11
#> 12 12
#> 13 13
#> 14 14
#> 15 15
#> 16 16
#> 17 17
#> 18 18
#> 19 19
#> 20 20
#> 21 21
#> 22 22
#> 23 23
#> 24 24
#> 25 25
#> 26 26
#> 27 27
Projecting time-specific niche models
To project the models, we use the predict method. Here, we will project one of the selected models using the environmental layers of 2016. Also, we project it using layers from a period that comprehends 1970-2000.
env_layers_2016 <- list.dirs(tempora_layers_dir,
recursive = FALSE)[32]
suit_2016 <- predict(mod_sel,
model_variables = c("bio_01","bio_03","bio_12"),
layers_path =env_layers_2016 ,
layers_ext = ".tif$")
#> | | | 0% | |======================================================================| 100%
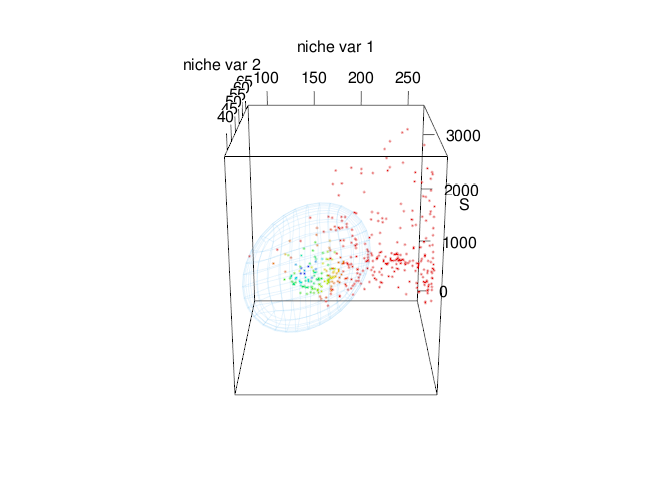
Fig. 3. A selected niche model projected using environmental layers from 2016.
Now for the period that comprehends 1970-2000.
layers_70_00_dir <- system.file("extdata/bio_1970_2000",package = "tenm")
suit_1970_2000 <- predict(mod_sel,
model_variables = c("bio_01","bio_03","bio_12"),
layers_path = layers_70_00_dir,
layers_ext = ".tif$")
#> | | | 0% | |======================================================================| 100%
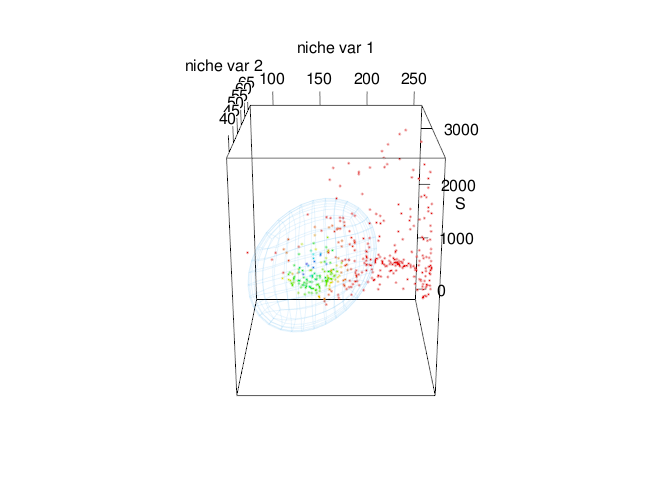
Fig. 4. A selected niche model projected using environmental layers from 1970-2000.
Lets see the predictions in geographic space
par(mfrow=c(1,2), mar=c(4,4,2,2))
terra::plot(suit_2016, main="Prediction for 2016")
terra::plot(suit_1970_2000, main="Prediction for 1970-2000")
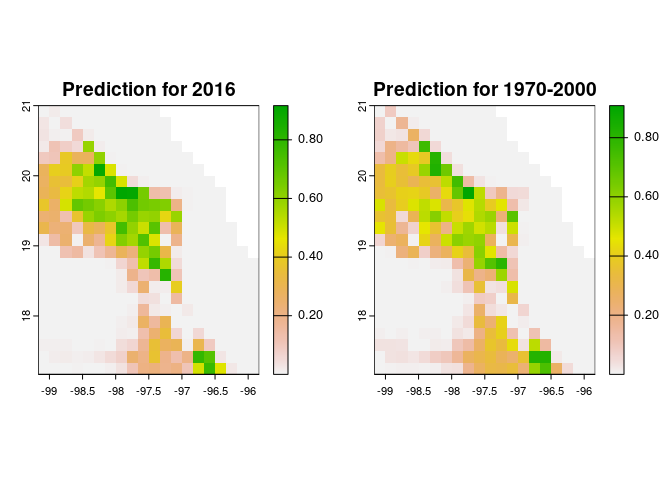
Fig. 5. Geographic projection of a selected model. Left panel, the projection using environmental layers from 2016. Right panel, the projection using environmental layers from 1970-2000
Comparing time-specific niche model vs. standard niche model
The following lines of code show the differences of a time-specific niche model and a standard niche model.
layers_70_00_dir <- system.file("extdata/bio_1970_2000",package = "tenm")
layers_70_00_path <- list.files(layers_70_00_dir,
pattern = ".tif$",full.names = TRUE)
# Extract environmental information
elayers_70_00 <- terra::rast(layers_70_00_path)
e_trad <- terra::extract(elayers_70_00,
ab_1[,c("decimalLongitude","decimalLatitude")])
rgl::view3d(theta = 0, phi = -60,fov=120, zoom = 0.7)
tenm::plot_ellipsoid(x = e_trad$bio_01,y=e_trad$bio_03,z=e_trad$bio_12,
col = "#1B9E77",
xlab = "Bio 1",
ylab = "Bio 3",
zlab = "Bio 12",)
tenm::plot_ellipsoid(x = abbg$temporal_df$bio_01,
y = abbg$temporal_df$bio_03,
z = abbg$temporal_df$bio_12,
col = "#E7298A",
add = TRUE)
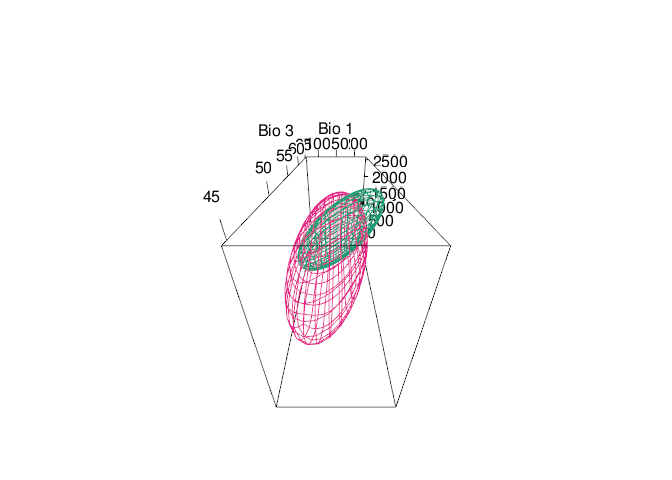
Fig. 6. Time-specific niche model vs. standard niche model. Pink ellipsoid represents the time-specific niche model. Green ellipsoid represents a ellipsoid model fitted using the standard approach.
Note that both ellipsoids differ in size and shape. In standard approach (green ellipsoid), we can see an sub-estimation of the environmental values where the intrinsic growth rate might be positive.
Acknowledgments
CONACYT Ciencia de Frontera CF-2023-I-1156. Laboratorio Nacional Conahcyt de Biología del Cambio Climático, México. To PAPIIT-UNAM IA202824 and PAPIIT-UNAM IA203922. RGCD thanks the Universidad Nacional Autónoma de México (Dirección General de Asuntos del Personal Académico, DGAPA-UNAM, México) for her postdoctoral scholarship.