Description
Calculate Wood Volumes from Taper Functions.
Description
Functions for estimation of wood volumes, number of logs, diameters along the stem and heights at which certain diameters occur, based on taper functions and other parameters. References: McTague, J. P., & Weiskittel, A. (2021). <doi:10.1139/cjfr-2020-0326>.
README.md
timbeR
The goal of timbeR is to provide functions for estimating log volumes and quantities from taper functions in the processing of forest inventories.
Installation
You can install the CRAN version of the package as follows:
install.packages("timbeR")
You can also install the development version from GitHub with:
# install.packages("devtools")
devtools::install_github("sergiocostafh/timbeR")
Example
As a basic example, we can fit a 5th degree polynomial to the tree_scaling dataset.
library(dplyr)
library(timbeR)
tree_scaling <- tree_scaling %>%
mutate(did = di/dbh,
hih = hi/h)
poli5 <- lm(did~hih+I(hih^2)+I(hih^3)+I(hih^4)+I(hih^5),tree_scaling)
And then we define the wood products:
assortments <- data.frame(
NAME = c('15-25','4-15'),
SED = c(15,4),
MINLENGTH = c(2.65,2),
MAXLENGTH = c(2.65,4.2),
LOSS = c(5,5)
)
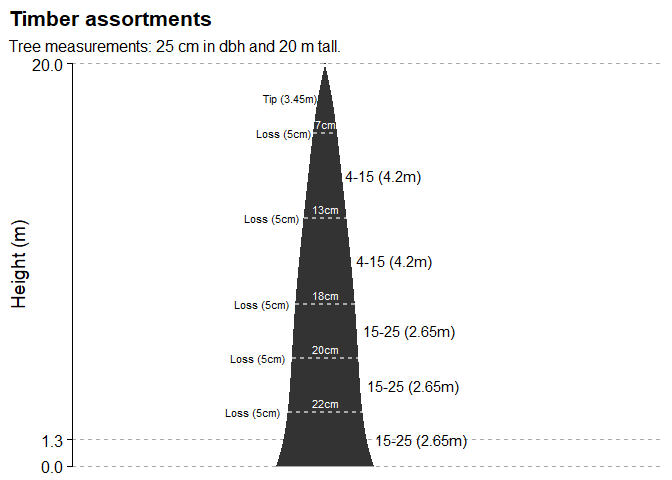
And now we can estimate the volume and quantity of wood products in a tree stem. For ease of understanding, let’s simulate cutting logs on a single tree.
# Tree measurements
dbh <- 25
h <- 20
# Estimate logs volume and quantity
poly5_logs(dbh, h, coef(poli5), assortments)
#> $volumes
#> # A tibble: 1 x 2
#> `15-25` `4-15`
#> <dbl> <dbl>
#> 1 0.293 0.111
#>
#> $logs
#> # A tibble: 1 x 2
#> `15-25` `4-15`
#> <dbl> <dbl>
#> 1 3 2
Finally, we can generate the same result in a visual way, simulating the position of the logs along the tree stem.
poly5_logs_plot(dbh, h, coef(poli5), assortments)