Generate Graphviz documents from a Haskell representation.
Render graphs using a declarative markup. Currently supports DFD (http:/en.wikipedia.orgwiki/Data_flow_diagram) and sequence diagrams (http:/plantuml.sourceforge.netsequence.html).
DataFlow
Render graphs using a declarative markup. Currently supports DFD (http://en.wikipedia.org/wiki/Data_flow_diagram) and sequence diagrams (http://plantuml.sourceforge.net/sequence.html).
Usage
The following forms are supported by DataFlow.
IDs
An ID can contain letters, numbers and underscores. It must start with a letter.
my_id_contain_4_words
Strings
String literals are written using double quotes.
"this is a string and it can contain everything but double quotes and newlines"
NOTE! Escaping characters inside strings is not supported at the moment.
Text Blocks
Text blocks are special strings, enclosed in backticks, that are can span multiple lines in the source document. The space characters before the first non-space characters on each line are trimmed, regardless of the indentation.
`this is
a
textblock`
... is converted to:
this is
a
textblock
Arrays
Arrays can contain other values (arrays, strings, text blocks).
["hello", "world", ["I", "am", `nested
here`]]
Attributes
Attributes are key-value pairs for diagrams and nodes that are used by output renderers. Attributes are enclosed by curly brackets. For nodes that can contain other nodes, attributes must appear before nodes.
Keys have the same rules as IDs. Values can be strings or text blocks.
{
key1 = "attr value"
key2 = `attr
value`
key3 = ["value1", "value2"]
}
diagram
diagram is the top-level form and must appear exactly once in a DataFlow document. It can contain attributes and nodes.
diagram {
title = "My diagram"
}
boundary
The boundary form declares a TrustBoundary node that can contain attributes and other nodes. Boundaries are only allowed in the top-level diagram and they must have unique IDs.
diagram {
boundary my_boundary {
title = "My System"
}
}
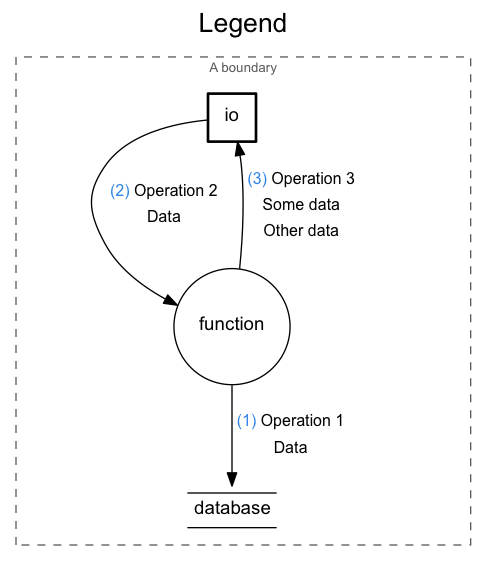
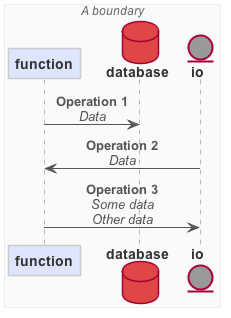
nodes: io, function, database
The io, function and database forms declare InputOutput, Function and Database nodes, respectively. The nodes have IDs and they can contain attributes. Empty attribute brackets can be omitted.
diagram {
io thing1
io thing2 {
title = "Thing 2"
}
}
->
The -> form declares a Flow between the nodes referenced by their IDs. It can contain attributes. Empty attribute brackets can be omitted. Flows must be declared after all nodes.
Note that the arrow can be reversed as well (<-).
diagram {
thing1 -> thing2
thing1 <- thing2 {
operation = "Greet"
data = "A nice greeting"
}
}
Comment
Comments are written using /* and */ and are ignored by the Reader. They're only used for human consumption.
diagram {
/* I can write
* whatever I
* want in here! */
}
Example
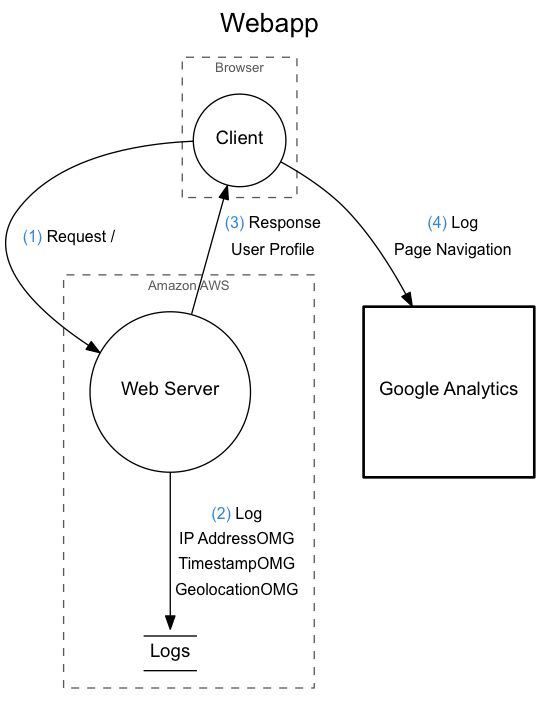
The image from the top of this README is rendered from the following DataFlow document.
diagram {
title = "Webapp"
/* Some comment about this... */
threats = `
No particular threats at this point.
It's **extremely** safe.`
boundary {
title = "Browser"
function client {
title = "Client"
}
}
boundary {
title = "Amazon AWS"
function server {
title = "Web Server"
}
database logs {
title = "Logs"
}
}
io analytics {
title = "Google Analytics"
}
client -> server {
operation = "Request /"
description = `User navigates with a browser to see some content.`
}
server -> logs {
operation = "Log"
data = `The user
IP address.`
description = `Logged to a ELK stack.`
}
server -> client {
operation = "Response"
data = "User Profile"
description = `The server responds with some HTML.`
}
analytics <- client {
operation = "Log"
data = "Page Navigation"
description = `The Google Analytics plugin sends navigation
data to Google.`
}
}
Run DataFlow
The dataflow executable takes an output format and a DataFlow source document and writes the output to stdout.
dataflow (dfd|seq) FILE
DFD
To use the DFD output you need Graphviz installed.
dataflow dfd webapp.flow | dot -Tpng > webapp.png
Output

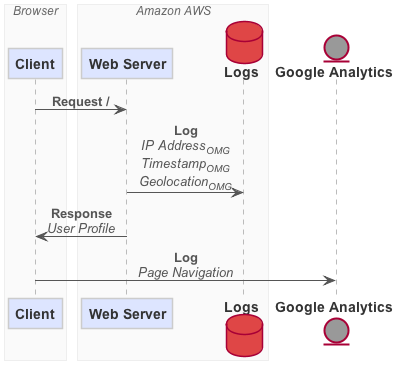
Sequence Diagram
You can use PlantUML to generate a sequence diagram.
dataflow seq webapp.flow | java -Djava.awt.headless=true -jar plantuml.jar -tpng -pipe > webapp.png
Output
Templating
You can use Hastache to output arbitrary text with its Mustache-like templates.
dataflow template template.ha webapp.flow > webapp.html
Built-in Functions and Values
markdown- Convert the attribute at the given key from Markdown to HTML.{{#markdown}}my_markdown_attr{{/markdown}}html_linebreaks- Replace\nwith<br/>elements in the attribute at the given key, to retain linebreaks in HTML output.{{#html_linebreaks}}my_formatted_attr{{/html_linebreaks}}filename_without_extension- The input.flowfile name with no path and no extension. Useful when generating graphics and text/HTML with matching filenames (e.g.my-flow.htmlincludesmy-flow.png).<img src="{{filename_without_extension}}.png" />flows- a list of all the Flow nodes in the diagram. Attributes of the flow is accessible inside the iteration scope, including anumber.<ol> {{#flows}} <li>{{number}} - {{description}}</li> {{/flows}} </ol>
For an example see template.ha and the output HTML in webapp.html.
Output

Makefile Example
The following Makefile finds .flow sources in src and generates DFDs, in SVG format, in dist.
SOURCES=$(shell find src/*.flow)
TARGETS=$(SOURCES:src/%.flow=dist/%.dfd.svg)
K := $(if $(shell which dataflow),,$(error "No dataflow executable in PATH. See https://github.com/SonyMobile/dataflow for install instructions)))"))
dist/%.dfd.svg: src/%.flow
@dataflow dfd $< | dot -Tsvg > $@
dfd: $(TARGETS)
clean:
rm -f $(TARGETS)
Build Instructions
See BUILD.md.
License
BSD-3, see LICENSE.