Biotic Ligand Model Engine.
Biotic Ligand Model Engine in R
THIS PACKAGE IS A WORK IN PROGRESS. This package is the Biotic Ligand Model (BLM) engine developed by Windward Environmental, LLC. The BLM was originally programmed in 'PowerBasic' by Robert Santore and others. The main way the BLM can be used is to predict the toxicity of a metal to an organism with a known sensitivity (i.e., it is known how much of that metal must accumulate on that organism's biotic ligand to cause a physiological effect in a certain percentage of the population, such as a 20% loss in reproduction or a 50% mortality rate). The second way the BLM can be used is to estimate the chemical speciation of water, including estimating the amount of metal accumulated to an organism's biotic ligand during a toxicity test. In the first application of the BLM, the amount of metal will be predicted, while in the second application, the amount of metal is known and the portions of that metal that exist in various forms will be determined.
Overview
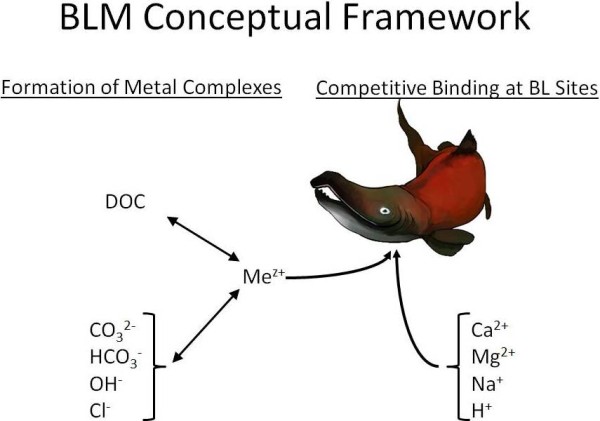
The bioavailability of metals in aquatic systems (i.e., the amount of metal that is available for uptake by organisms) is highly dependent on several factors. For example, dissolved organic carbon (DOC) in the water binds metals, cations (such as calcium) can compete with metals for uptake by an organism, and pH and alkalinity affect metal speciation (Figure 1). For a metal like copper, bioavailability is greatest in waters with low DOC, low hardness, and low pH; bioavailability generally decreases as any one of the parameters increases. Other metals, such as aluminum, have more complex speciation.
 {alt="BLM Conceptual Framework"}
{alt="BLM Conceptual Framework"}
For example, highly bioavailable forms of aluminum can occur at both pH 6 and 8, but less bioavailable forms occur at pH 7. The biotic ligand model (BLM) is a tool that can mechanistically predict the bioavailability of a variety of metals under the large range of water chemistry conditions that are observed in nature. The BLM is scientifically robust and defensible, user friendly, and freely available for download from this website.
Examples of BLM Applications
The US Environmental Protection Agency’s (EPA’s) recommended freshwater ambient water quality criteria (AWQC) for copper are based on the BLM.
- BLM-based saltwater AWQC for copper in saltwater are pending.
BLM-based, environmentally protective levels for several other metals have been developed or are currently in development (e.g., aluminum, cadmium, lead, nickel, and zinc).
Site-specific water quality criteria can be developed for use in determining effluent permit limits, or in state-wide water quality assessments required under Section 305(b) of the Clean Water Act.
The BLM can be used to support site-specific ecological risk assessments.
For toxicity identification evaluations (TIEs), the BLM can evaluate potential causes of toxicity in whole effluent toxicity (WET) tests.
The BLM can also be used to evaluate the bioavailability of metal mixtures.
Current BLMs
Freshwater: aluminum, cadmium, cobalt, copper, nickel, lead, and zinc
Saltwater: copper, nickel, and zinc
Contact Information
For questions or problems relating to the application of the BLM, please contact:
Robert C. Santore
Windward Environmental
e-mail Robert
Phone: (206) 812-5450
Relevant Literature
Santore R.C., Ryan A.C., Kroglund F., Rodriguez P., Stubblefield W., Cardwell A., Adams W., Nordheim E. 2018. Development and application of a biotic ligand model for predicting the chronic toxicity of dissolved and precipitated aluminum to aquatic organisms. Environmental Toxicology & Chemistry 37(1):70-79. https://doi.org/10.1002/etc.4020
DeForest D.K., Santore R.C., Ryan A.C., Church B., Chowdhury J.M., Brix K.V.. 2017. Development of biotic ligand model (BLM)-based freshwater aquatic life criteria for lead following U.S. EPA guidelines. Environmental Toxicology & Chemistry 36(11):2965-2973. https://doi.org/10.1002/etc.3861
Santore R.C., Ryan A.C.. 2015. Development and application of a multi-metal multi-biotic ligand model for assessing aquatic toxicity of metal mixtures. Environmental Toxicology & Chemistry 34:777-787. https://doi.org/10.1002/etc.2869
Bosse C., Rosen G., Colvin M., Earley P., Santore R., Rivera-Duarte I. 2014. Copper bioavailability and toxicity to Mytilus galloprovincialis in Shelter Island Yacht Basin, San Diego, CA. Marine Pollution Bulletin 85:225-234. https://doi.org/10.1016/j.marpolbul.2014.05.045
DeForest D.K., Van Genderen E.J. 2012. Application of USEPA guidelines in a bioavailability-based assessment of ambient water quality criteria for zinc in freshwater. Environmental Toxicology & Chemistry 31:1264-1272. https://doi.org/10.1002/etc.1810
Ryan A.C., Tomasso J.R., Klaine S.J.. 2009. Influence of pH, hardness, dissolved organic carbon concentration, and dissolved organic matter source on the acute toxicity of copper to Daphnia magna in soft waters: implications for the biotic ligand model. Environmental Toxicology & Chemistry 28:1663-1670. https://doi.org/10.1897/08-361.1
Bielmyer G.K., Grosell M., Paquin P.R., Mathews R., Wu K.B., Santore R.C., Brix K.V. 2007. Validation study of the acute biotic ligand model for silver. Environmental Toxicology & Chemistry 26:2241-2246. https://doi.org/10.1897/06-634R.1
Arnold W.R., Santore R.C., Cotsifas J.S. 2005. Predicting copper toxicity in estuarine and marine waters using the Biotic Ligand Model. Marine Pollution Bulletin 50:1634-1640. https://doi.org/10.1016/j.marpolbul.2005.06.035
DiToro D.M., McGrath J.A., Hansen D.J., Berry W.J., Paquin P.R., Mathew R., Wu K.B., Santore R.C. 2005. Predicting sediment metal toxicity using a sediment biotic ligand model: methodology and initial application. Environmental Toxicology & Chemistry 24(10):2410-2427. https://doi.org/10.1897/04-413R.1
Ryan A.C., Van Genderen E.J., Tomasso J.R., Klaine S.J. 2004. Influence of natural organic matter source on copper toxicity to larval fathead minnows (Pimephales promelas): implications for the biotic ligand model. Environmental Toxicology & Chemistry 23:1567-1574. https://doi.org/10.1897/02-476
Santore R.C., Mathew R., Paquin P.R., DiToro D.M.. 2002. Application of the biotic ligand model to predicting zinc toxicity to rainbow trout, fathead minnow, and Daphnia magna. Aquatic Toxicology 133:271-285. https://doi.org/10.1016/S1532-0456(02)00106-0{.uri}
Paquin P.R., Gorsuch J.W., Apte S., Batley G.E., Bowles K.C., Campbell P.G.C., Delos C.G., DiToro D.M., Dwyer R.L., Galvez F., Gensemer R.W., Goss G.G., Hogstrand C., Janssen C.R., McGeer J.C., Naddy R.B., Playle R.C., Santore R.C., Schneider U., Stubblefield W.A., Wood C.M., Wu K.B. 2002. The biotic ligand model: a historical overview. Comparative Biochemistry & Physiology Part C 133:3-35. https://doi.org/10.1016/S1532-0456(02)00112-6{.uri}
DiToro D.M., Allen H.E., Bergman H.L., Meyer J.S., Paquin P.R., Santore R.C. 2001. Biotic ligand model of the acute toxicity of metals. 1. Technical basis. Environmental Toxicology & Chemistry 20:2383-2396. https://doi.org/10.1002/etc.5620201034
Santore R.C., DiToro D.M., Paquin P.R., Allen H.E., Meyer J.S. 2001. Biotic ligand model of the acute toxicity of metals. 2. Application to acute copper toxicity in freshwater fish and Daphnia. Environmental Toxicology & Chemistry 20:2397-2402. https://doi.org/10.1002/etc.5620201035