Biterm Topic Models for Short Text.
BTM - Biterm Topic Modelling for Short Text with R
This is an R package wrapping the C++ code available at https://github.com/xiaohuiyan/BTM for constructing a Biterm Topic Model (BTM). This model models word-word co-occurrences patterns (e.g., biterms).
Topic modelling using biterms is particularly good for finding topics in short texts (as occurs in short survey answers or twitter data).
Installation
This R package is on CRAN, just install it with install.packages('BTM')
What
The Biterm Topic Model (BTM) is a word co-occurrence based topic model that learns topics by modeling word-word co-occurrences patterns (e.g., biterms)
- A biterm consists of two words co-occurring in the same context, for example, in the same short text window.
- BTM models the biterm occurrences in a corpus (unlike LDA models which model the word occurrences in a document).
- It's a generative model. In the generation procedure, a biterm is generated by drawing two words independently from a same topic
z. In other words, the distribution of a bitermb=(wi,wj)is defined as:P(b) = sum_k{P(wi|z)*P(wj|z)*P(z)}where k is the number of topics you want to extract. - Estimation of the topic model is done with the Gibbs sampling algorithm. Where estimates are provided for
P(w|k)=phiandP(z)=theta.
More detail can be referred to the following paper:
Xiaohui Yan, Jiafeng Guo, Yanyan Lan, Xueqi Cheng. A Biterm Topic Model For Short Text. WWW2013. https://github.com/xiaohuiyan/xiaohuiyan.github.io/blob/master/paper/BTM-WWW13.pdf
Example
library(udpipe)
library(BTM)
data("brussels_reviews_anno", package = "udpipe")
## Taking only nouns of Dutch data
x <- subset(brussels_reviews_anno, language == "nl")
x <- subset(x, xpos %in% c("NN", "NNP", "NNS"))
x <- x[, c("doc_id", "lemma")]
## Building the model
set.seed(321)
model <- BTM(x, k = 3, beta = 0.01, iter = 1000, trace = 100)
## Inspect the model - topic frequency + conditional term probabilities
model$theta
[1] 0.3406998 0.2413721 0.4179281
topicterms <- terms(model, top_n = 10)
topicterms
[[1]]
token probability
1 appartement 0.06168297
2 brussel 0.04057012
3 kamer 0.02372442
4 centrum 0.01550855
5 locatie 0.01547671
6 stad 0.01229227
7 buurt 0.01181460
8 verblijf 0.01155985
9 huis 0.01111402
10 dag 0.01041345
[[2]]
token probability
1 appartement 0.05687312
2 brussel 0.01888307
3 buurt 0.01883812
4 kamer 0.01465696
5 verblijf 0.01339812
6 badkamer 0.01285862
7 slaapkamer 0.01276870
8 dag 0.01213928
9 bed 0.01195945
10 raam 0.01164474
[[3]]
token probability
1 appartement 0.061804812
2 brussel 0.035873377
3 centrum 0.022193831
4 huis 0.020091282
5 buurt 0.019935537
6 verblijf 0.018611710
7 aanrader 0.014614272
8 kamer 0.011447470
9 locatie 0.010902365
10 keuken 0.009448751
scores <- predict(model, newdata = x)
Make a specific topic called the background
# If you set background to TRUE
# The first topic is set to a background topic that equals to the empirical word distribution.
# This can be used to filter out common words.
set.seed(321)
model <- BTM(x, k = 5, beta = 0.01, background = TRUE, iter = 1000, trace = 100)
topicterms <- terms(model, top_n = 5)
topicterms
Visualisation of your model
- Can be done using the textplot package (https://github.com/bnosac/textplot), which can be found at CRAN as well (https://cran.r-project.org/package=textplot)
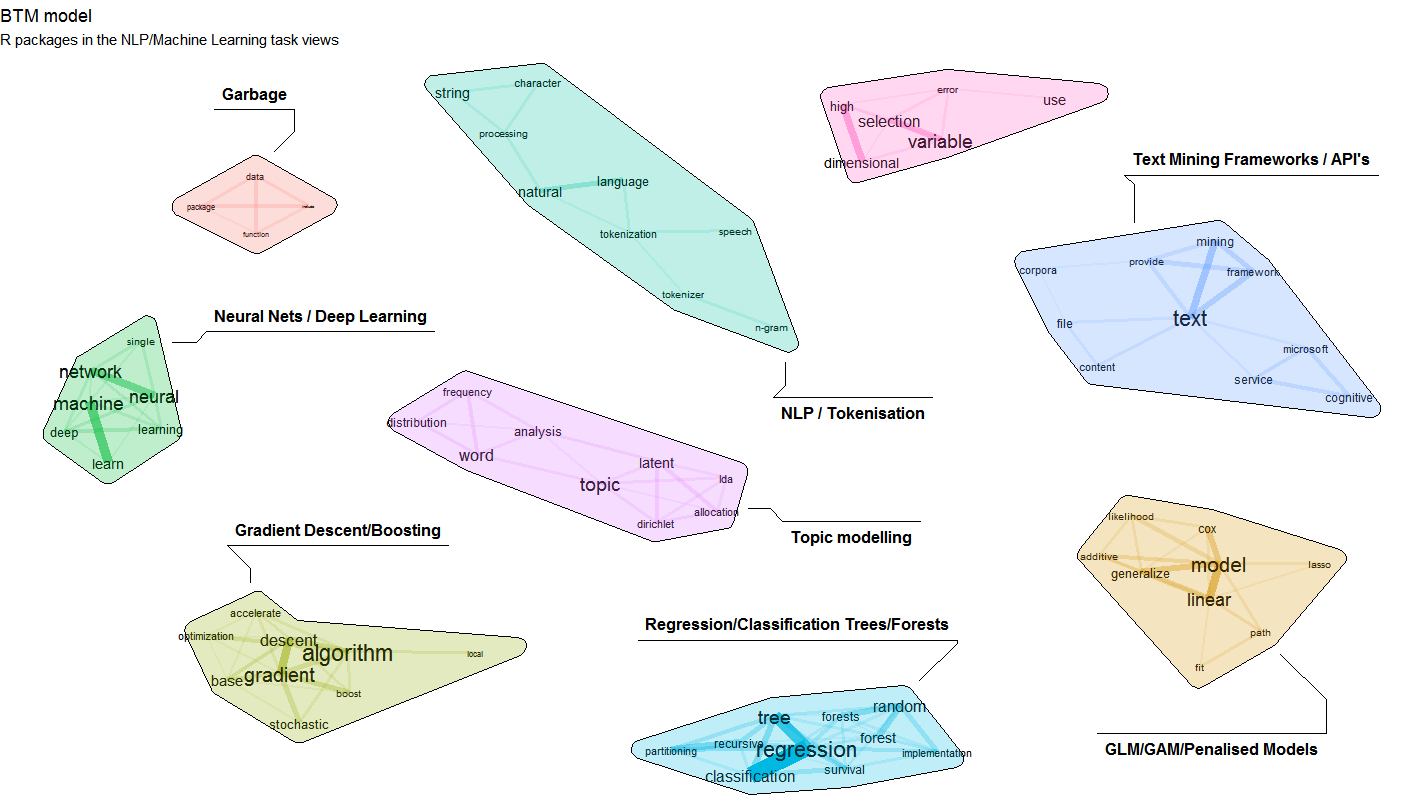
- An example visualisation built on a model of all R packages from the Natural Language Processing and Machine Learning task views is shown above (see also https://www.bnosac.be/index.php/blog/98-biterm-topic-modelling-for-short-texts)
library(textplot)
library(ggraph)
library(concaveman)
plot(model)
Provide your own set of biterms
An interesting use case of this package is to
- cluster based on parts of speech tags like nouns and adjectives which can be found in the text in the neighbourhood of one another
- cluster dependency relationships provided by NLP tools like udpipe (https://CRAN.R-project.org/package=udpipe)
This can be done by providing your own set of biterms to cluster upon.
Example clustering cooccurrences of nouns/adjectives
library(data.table)
library(udpipe)
## Annotate text with parts of speech tags
data("brussels_reviews", package = "udpipe")
anno <- subset(brussels_reviews, language %in% "nl")
anno <- data.frame(doc_id = anno$id, text = anno$feedback, stringsAsFactors = FALSE)
anno <- udpipe(anno, "dutch", trace = 10)
## Get cooccurrences of nouns / adjectives and proper nouns
biterms <- as.data.table(anno)
biterms <- biterms[, cooccurrence(x = lemma,
relevant = upos %in% c("NOUN", "PROPN", "ADJ"),
skipgram = 2),
by = list(doc_id)]
## Build the model
set.seed(123456)
x <- subset(anno, upos %in% c("NOUN", "PROPN", "ADJ"))
x <- x[, c("doc_id", "lemma")]
model <- BTM(x, k = 5, beta = 0.01, iter = 2000, background = TRUE,
biterms = biterms, trace = 100)
topicterms <- terms(model, top_n = 5)
topicterms
Example clustering dependency relationships
library(udpipe)
library(tm)
library(data.table)
data("brussels_reviews", package = "udpipe")
exclude <- stopwords("nl")
## Do annotation on Dutch text
anno <- subset(brussels_reviews, language %in% "nl")
anno <- data.frame(doc_id = anno$id, text = anno$feedback, stringsAsFactors = FALSE)
anno <- udpipe(anno, "dutch", trace = 10)
anno <- setDT(anno)
anno <- merge(anno, anno,
by.x = c("doc_id", "paragraph_id", "sentence_id", "head_token_id"),
by.y = c("doc_id", "paragraph_id", "sentence_id", "token_id"),
all.x = TRUE, all.y = FALSE, suffixes = c("", "_parent"), sort = FALSE)
## Specify a set of relationships you are interested in (e.g. objects of a verb)
anno$relevant <- anno$dep_rel %in% c("obj") & !is.na(anno$lemma_parent)
biterms <- subset(anno, relevant == TRUE)
biterms <- data.frame(doc_id = biterms$doc_id,
term1 = biterms$lemma,
term2 = biterms$lemma_parent,
cooc = 1,
stringsAsFactors = FALSE)
biterms <- subset(biterms, !term1 %in% exclude & !term2 %in% exclude)
## Put in x only terms whch were used in the biterms object such that frequency stats of terms can be computed in BTM
anno <- anno[, keep := relevant | (token_id %in% head_token_id[relevant == TRUE]), by = list(doc_id, paragraph_id, sentence_id)]
x <- subset(anno, keep == TRUE, select = c("doc_id", "lemma"))
x <- subset(x, !lemma %in% exclude)
## Build the topic model
model <- BTM(data = x,
biterms = biterms,
k = 6, iter = 2000, background = FALSE, trace = 100)
topicterms <- terms(model, top_n = 5)
topicterms
Support in text mining
Need support in text mining? Contact BNOSAC: http://www.bnosac.be.