Composite Grid Gaussian Processes.
CGGP
The goal of CGGP is to provide a sequential design of experiment algorithm that can efficiently use many points and interpolate exactly.
Installation
You can install CGGP from GitHub with:
# install.packages("devtools")
devtools::install_github("CollinErickson/CGGP")
Example
To create a CGGP object:
## basic example code
library(CGGP)
d <- 4
CG <- CGGPcreate(d=d,200)
print(CG)
#> CGGP object
#> d = 4
#> output dimensions = 1
#> CorrFunc = PowerExponential
#> number of design points = 193
#> number of unevaluated design points = 193
#> Available functions:
#> - CGGPfit(CGGP, Y) to update parameters with new data
#> - CGGPpred(CGGP, xp) to predict at new points
#> - CGGPappend(CGGP, batchsize) to add new design points
#> - CGGPplot<name>(CGGP) to visualize CGGP model
A new CGGP object has design points that should be evaluated next, either from CG$design or CG$design_unevaluated.
f <- function(x) {x[1]^2*cos(x[3]) + 4*(0.5-x[2])^3*(1-x[1]/3) + x[1]*sin(2*2*pi*x[3]^2)}
Y <- apply(CG$design, 1, f)
Once you have evaluated the design points, you can fit the object with CGGPfit.
CG <- CGGPfit(CG, Y)
CG
#> CGGP object
#> d = 4
#> output dimensions = 1
#> CorrFunc = PowerExponential
#> number of design points = 193
#> number of unevaluated design points = 0
#> Available functions:
#> - CGGPfit(CGGP, Y) to update parameters with new data
#> - CGGPpred(CGGP, xp) to predict at new points
#> - CGGPappend(CGGP, batchsize) to add new design points
#> - CGGPplot<name>(CGGP) to visualize CGGP model
If you want to use the model to make predictions at new input points, you can use CGGPpred.
xp <- matrix(runif(10*CG$d), ncol=CG$d)
CGGPpred(CG, xp)
#> $mean
#> [,1]
#> [1,] -0.52367139
#> [2,] -0.31521070
#> [3,] 0.05781108
#> [4,] 0.94199482
#> [5,] 0.98578179
#> [6,] -0.22661334
#> [7,] 0.17936738
#> [8,] -0.16986576
#> [9,] 0.73173845
#> [10,] -0.15488181
#>
#> $var
#> [,1]
#> [1,] 0.0092572099
#> [2,] 0.0221140651
#> [3,] 0.0094446082
#> [4,] 0.0134280357
#> [5,] 0.0012771995
#> [6,] 0.0002103327
#> [7,] 0.0035323769
#> [8,] 0.0149844852
#> [9,] 0.0170830979
#> [10,] 0.0183621344
To add new design points to the already existing design, use CGGPappend. It will use the data already collected to find the most useful set of points to evaluate next.
# To add 100 points
CG <- CGGPappend(CG, 100)
Now you will need to evaluate the points added to CG$design, and refit the model.
ynew <- apply(CG$design_unevaluated, 1, f)
CG <- CGGPfit(CG, Ynew=ynew)
Plot functions
There are a few functions that will help visualize the CGGP design.
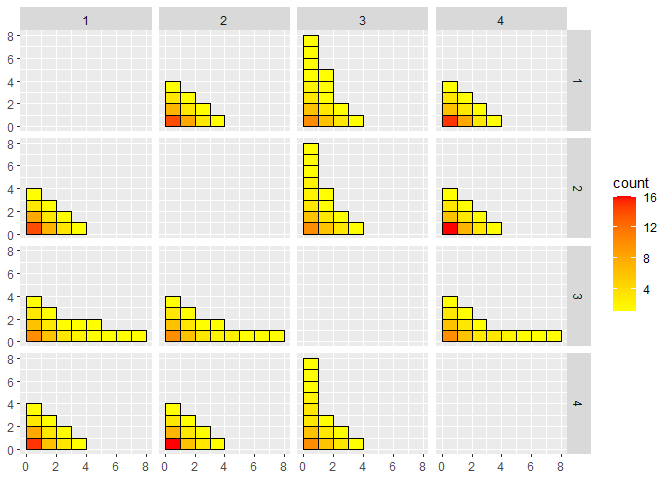
CGGPplotblocks
CGGPplotblocks shows the block structure when projected down to all pairs of two dimensions. The plot is symmetric. The facet labels be a little bit confusing. The first column has the label 1, and it looks like it is saying that the x-axis for each plot in that column is for X1, but it is actually the y-axis that is X1 for each plot in that column.
CGGPplotblocks(CG)
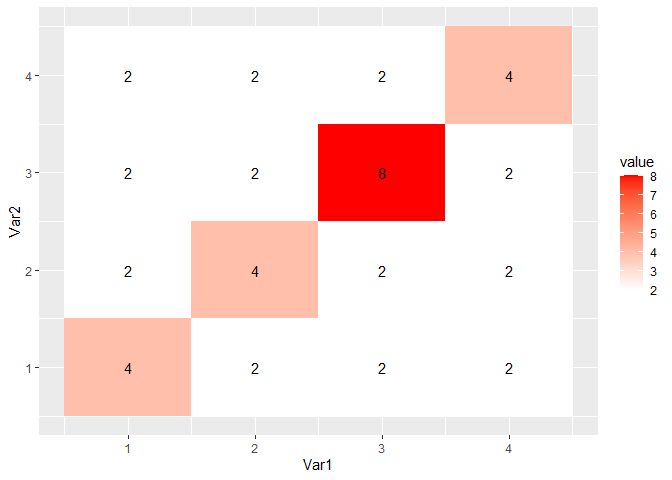
CGGPplotheat
CGGPplotheat is similar to CGGPplotblocks and can be easier to read since it is only a single plot. The $(i,j)$ entry shows the maximum value for which a block was selected with $X_i$ and $X_j$ at least that large. The diagonal entries, $(i, i)$, show the maximum depth for $X_i$. A diagonal entry must be at least as large as any entry in its column or row. This plot is also symmetric.
CGGPplotheat(CG)
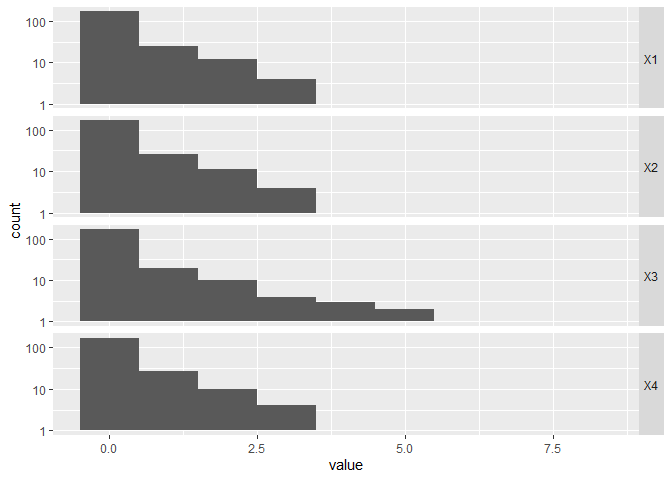
CGGPhist
CGGPhist shows histograms of the block depth in each direction. The dimensions that have more large values are dimensions that have been explored more. These should be the more active dimensions.
CGGPplothist(CG)
#> Warning: Transformation introduced infinite values in continuous y-axis
#> Warning: Removed 12 rows containing missing values (`geom_bar()`).
CGGPplotcorr
CGGPplotcorr gives an idea of what the correlation structure in each dimension is. The values plotted do not represent the actual data given to CGGP. Each wiggly line represents a random Gaussian process drawn using the correlation parameters for that dimension from the given CGGP model. Dimensions that are more wiggly and have higher variance are the more active dimensions. Dimensions with nearly flat lines mean that the corresponding input dimension has a relatively small effect on the output.
CGGPplotcorr(CG)
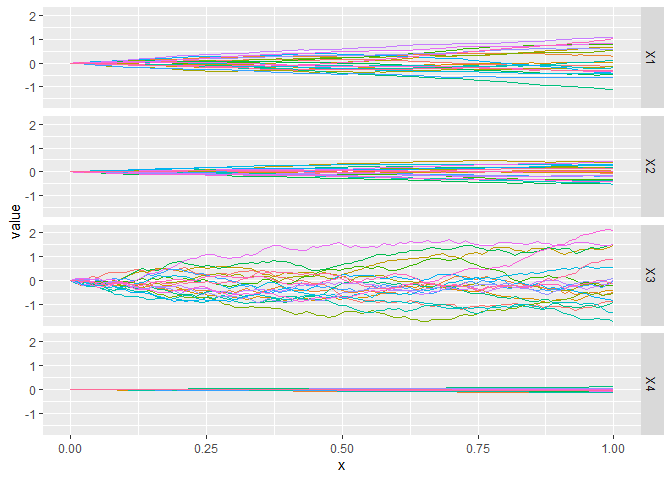
CGGPplotvariogram
CGGPplotvariogram shows something similar to the semi-variogram for the correlation parameters found for each dimension. Really it is just showing how the correlation function decays for points that are further away. It should always start at y=1 for x=0 and decrease in y as x gets larger
CGGPplotvariogram(CG)
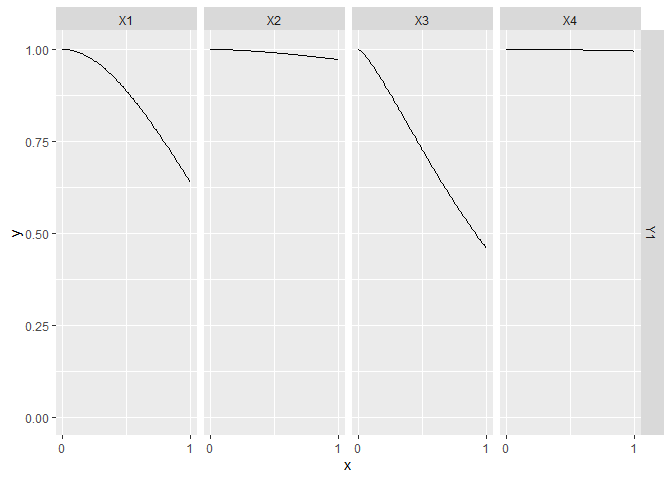
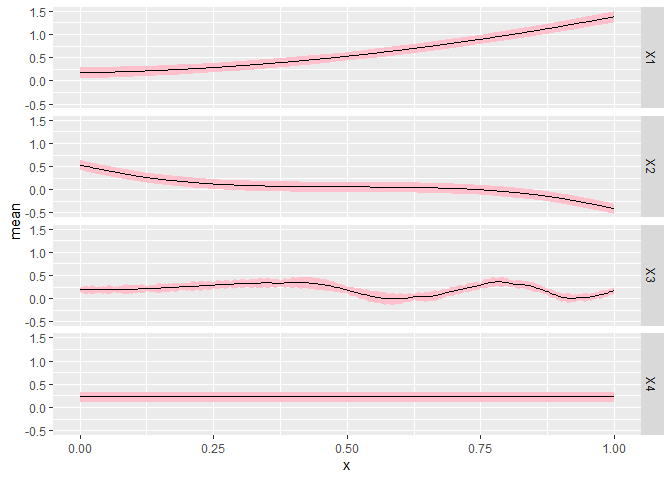
CGGPplotslice
CGGPplotslice shows what the predicted model along each individual dimension when the other input dimensions are held constant, i.e., a slice along a single dimension. By default the slice is done holding all other inputs at 0.5, but this can be changed by changing the argument proj. The black dots are the data points that are in that slice If you change proj to have values not equal to 0.5, you probably won’t see any black dots. The pink regions are the 95% prediction intervals.
This plot is the best for giving an idea of what the higher dimension function look like. You can see how the output changes as each input is varied.
CGGPplotslice(CG)
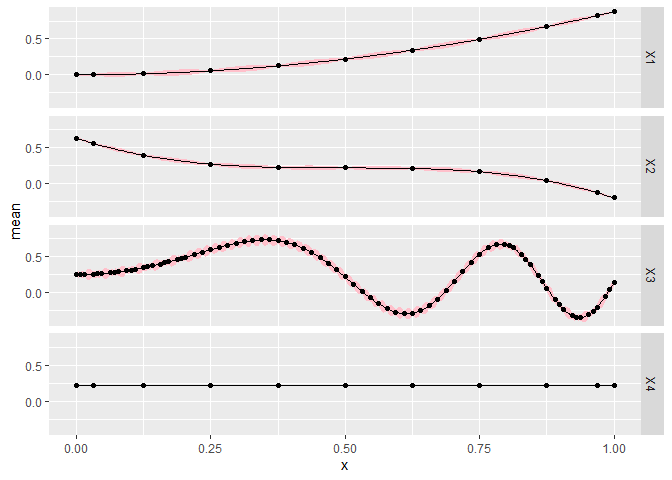
The next plot changes so that all the other dimensions are held constant at 0.15 for each slice plot. When moving from the center line, the error bounds generally should be larger since it is further from the data, but we should see similar patterns unless the function is highly nonlinear.
CGGPplotslice(CG, proj = rep(.15, CG$d))
CGGPplottheta
CGGPplottheta is useful for getting an idea of how the samples for the correlation parameters (theta) vary compared to the maximum a posteriori (MAP). This may be helpful when using UCB or TS in CGGPappend to get an idea of how much uncertainty there is in the parameters. Note that there are likely multiple parameters for each input dimension.
CGGPplottheta(CG)
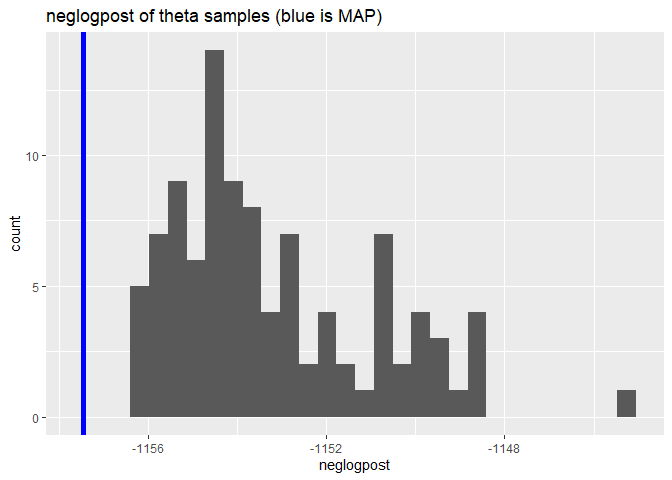
CGGPplotsamplesneglogpost
CGGPplotsamplesneglogpost shows the negative log posterior for each of the different samples for theta. The value for the MAP is shown as a blue line. It should be at the far left edge if it is the true MAP.
CGGPplotsamplesneglogpost(CG)