Plots the CHOIR Body Map.
CHOIRBM
CHOIRBM is a collection of plotting utilities for the Collaborative Health Outcomes Information Registry’s Body Map (CBM). The CBM is an instrument for assessing the distribution of a patient’s pain, and has been validated in a paper published in Pain Reports (Scherrer et al 2021). The package is built on top of the popular R plotting package ggplot2, and returns plots as ggplot objects.
Sample of the CBM in clinical use: 
Installation
You can install the released version of CHOIRBM from CRAN with: (Not on CRAN…yet)
install.packages("CHOIRBM")
Or install from GitHub with:
devtools::install_github("emcramer/CHOIRBM")
remotes::install_github("emcramer/CHOIRBM")
Examples
Plotting the Male CBM
This is a basic example which shows you how to plot the front and back parts of the male CHOIR Body Map:
library(CHOIRBM)
# generate some random example data
set.seed(123)
ids <- as.character(c(seq.int(101, 136, 1), seq.int(201, 238, 1)))
values <- data.frame(
id = ids
, value = runif(length(ids))
, ucolors = rainbow(length(ids))
, group = ifelse(as.numeric(ids) < 200, "Front", "Back")
)
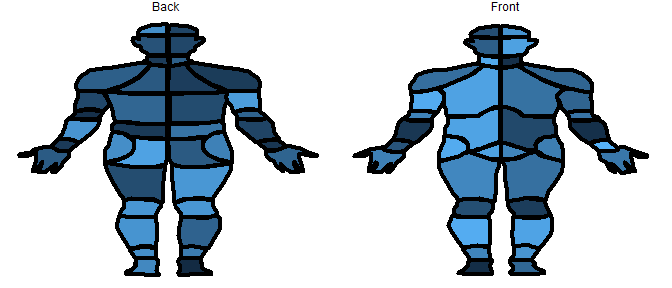
# plot the data on the front of the CHOIR body map
plot_male_choirbm(values, "value")
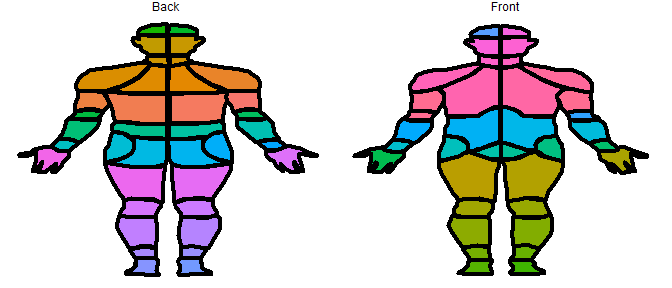
# plot each segment of the bodymap as a different color
plot_male_choirbm(values, "ucolors")
Plotting the female CBM
And an additional example with the female CHOIR Body Map:
library(CHOIRBM)
# generate some random example data
exdata <- gen_example_data()
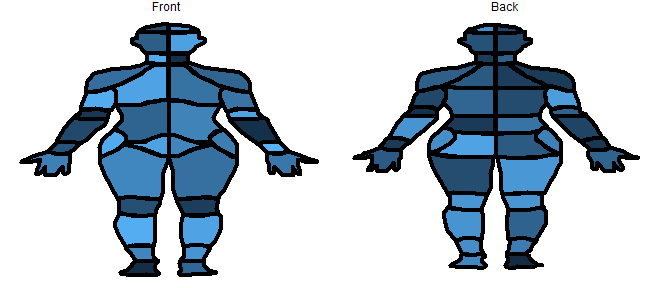
# plot the data on the front of the CHOIR body map
plot_female_choirbm(exdata, "value")
Check the vignettes for more examples such as plotting individual patients, highlighting specific segments, and changing color schemes.
Citations:
Scherrer, Kristen Hymel; Ziadni, Maisa S; Kong, Jiang-Tia; Sturgeon, John A; Salmasi, Vafia; Hong, Juliette; Cramer, Eric; Chen, Abby L; Pacht, Teresa; Olson, Garrick; Darnall, Beth D; Kao, Ming-Chih; Mackey, Sean. Development and validation of the Collaborative Health Outcomes Information Registry body map, PAIN Reports: January/February 2021 - Volume 6 - Issue 1 - p e880 doi: 10.1097/PR9.0000000000000880