Estimate the Confidence Intervals for Predictive Values.
Introduction
CIfinder is an R package intended to provide functions to compute confidence intervals for the positive predictive value (PPV) and negative predictive value (NPV) based on varied scenarios. In prospective studies where the proportion of diseased subjects provides an unbiased estimate of the disease prevalence, the confidence intervals for PPV and NPV can be estimated by methods that are applicable to single proportions. In this case, the package provides six methods for cocmputing the confidence intervals: “clopper.pearson”, “wald”, “wilson”, “wilson.correct”, “agresti”, and “beta”. In situations where the proportion of diseased subjects does not correspond to the disease prevalence (e.g. case-control studies), this package provides two types of solutions: I) three methods to estimate confidence intervals for PPV and NPV via ratio of two binomial proportions, including Gart & Nam (1988) https://doi.org/10.2307/2531848, Walter (1975) https://doi.org/10.1093/biomet/62.2.371, and MOVER-J (Laud, 2017) https://doi.org/10.1002/pst.1813; II) three direct methods to compute the confidence intervals, including Pepe (2003) Zhou (2007) <doi:10.1002/sim.2677, and Delta https://doi.org/10.1002/sim.2677. For more information, please see the Details and References sections in the user’s manual.
Installation
You can install the latest version of CIfinder by:
install.packages("CIfinder")
Example use
To demonstrate the utility of the CIfinder::ppv_npv_ci() function for calculating the confidence intervals for PPV and NPV, we will use a case-control study published by van de Vijver et al. (2002) https://doi.org/10.1056/NEJMoa021967. In the study, Cases were defined as those that have metastasis within 5 years of tumour excision, while controls were those that did not. Each tumor was classified as having a good or poor gene signature which was defined as having a signature correlation coefficient above or below the ‘optimized sensitivity’ threshold, respectively. This ‘optimized sensitivity’ threshold is defined as the correlation value that would result in a misclassification of at most 10 per cent of the cases. The performance of their 70-gene signature as prognosticator for metastasis is summarized as below:
#> Case Control
#> Poor_Signature 31 12
#> Good_Signature 3 32
#> Total 34 44
Generate the confidence interval for 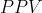 or
or 
Since this was a case-control study, the proportion of cases (34/(34+44)=43.6%) is not an unbiased estimate of its prevalence (assumed 7%). PPV and NPV and their confidence intervals can’t be estimated directly. To calculate the confidence interval based “gart and nam” method:
library(CIfinder)
ppv_npv_ci(x1 = 31, n1 = 34, x0 = 32, n0 = 44, prevalence = 0.07,
method = "gart and nam")
#> $method
#> [1] "gart and nam"
#>
#> $sensitivity
#> [1] 0.9117647
#>
#> $specificity
#> [1] 0.7272727
#>
#> $phi_ppv
#> phi_ppv_est phi_ppv_l phi_ppv_u phi_ppv_mle
#> 0.2991202 0.1715910 0.4656310 0.3009710
#>
#> $ppv
#> ppv_est ppv_l ppv_u ppv_mle
#> 0.2010444 0.1391548 0.3049051 0.2000554
#>
#> $phi_npv
#> phi_npv_est phi_npv_l phi_npv_u phi_npv_mle
#> 0.1213235 0.0323270 0.3090900 0.1266740
#>
#> $npv
#> npv_est npv_l npv_u npv_mle
#> 0.9909508 0.9772641 0.9975727 0.9905554
To calculate the confidence interval based “zhou’s method”
ppv_npv_ci(x1 = 31, n1 = 34, x0 = 32, n0 = 44, prevalence = 0.07,
method = "zhou")
#> $method
#> [1] "zhou"
#>
#> $sensitivity
#> [1] 0.9117647
#>
#> $specificity
#> [1] 0.7272727
#>
#> $ppv
#> ppv_est ppv_l ppv_u
#> 0.2010444 0.1217419 0.2803468
#>
#> $npv
#> npv_est npv_l npv_u
#> 0.9909508 0.9811265 1.0007750
#>
#> $ppv_logit_transformed
#> ppv_est ppv_l ppv_u
#> 0.2010444 0.1331384 0.2919216
#>
#> $npv_logit_transformed
#> npv_est npv_l npv_u
#> 0.9909508 0.9734141 0.9969560
In this output, since continuity correction isn’t specified, the estimates and confidence intervals in ppv and npv are same as the standard delta method where the ppv_logit_transformed and npv_logit_transformed refer the standard logit method described in the paper. If continuity.correction is being specified:
ppv_npv_ci(x1 = 31, n1 = 34, x0 = 32, n0 = 44, prevalence = 0.07,
method = "zhou",
continuity.correction = TRUE)
#> $method
#> [1] "zhou"
#>
#> $sensitivity
#> [1] 0.8699646
#>
#> $specificity
#> [1] 0.7090237
#>
#> $ppv
#> ppv_est ppv_l ppv_u
#> 0.1836999 0.1148465 0.2525533
#>
#> $npv
#> npv_est npv_l npv_u
#> 0.9863836 0.9750497 0.9977175
#>
#> $ppv_logit_transformed
#> ppv_est ppv_l ppv_u
#> 0.1836999 0.1244835 0.2626354
#>
#> $npv_logit_transformed
#> npv_est npv_l npv_u
#> 0.9863836 0.9688986 0.9940985
In this case, a continuity correction value is applied. The
ppv and npv outputs refer the “Adjusted” in the paper and ppv_logit_transformed and npv_logit_transformed denote the “Adjusted logit” method described in the paper.
Confidence interval for special cases
Assume we have a special case data where :
#> Case Control
#> Poor_Signature 31 0
#> Good_Signature 3 44
#> Total 34 44
In this situation, Pepe, Delta, and Zhou (standard logit) methods can not be used without continuity correction. Walter method can be used, but there may have skewness concerns. gart and nam and mover-j could be considered.
ppv_npv_ci(x1 = 31, n1 = 34, x0 = 44, n0 = 44, prevalence = 0.07,
method = "gart and nam")
#> $method
#> [1] "gart and nam"
#>
#> $sensitivity
#> [1] 0.9117647
#>
#> $specificity
#> [1] 1
#>
#> $phi_ppv
#> phi_ppv_est phi_ppv_l phi_ppv_u phi_ppv_mle
#> 0.000000 0.000000 0.067785 0.004121
#>
#> $ppv
#> ppv_est ppv_l ppv_u ppv_mle
#> 1.0000000 0.5261573 1.0000000 0.9480916
#>
#> $phi_npv
#> phi_npv_est phi_npv_l phi_npv_u phi_npv_mle
#> 0.08823529 0.02376800 0.21956500 0.09223300
#>
#> $npv
#> npv_est npv_l npv_u npv_mle
#> 0.9934025 0.9837423 0.9982142 0.9931056
Comparing to the Walter output:
ppv_npv_ci(x1 = 31, n1 = 34, x0 = 44, n0 = 44, prevalence = 0.07,
method = "walter")
#> $method
#> [1] "walter"
#>
#> $sensitivity
#> [1] 0.9117647
#>
#> $specificity
#> [1] 1
#>
#> $phi_ppv
#> phi lower upper
#> 0.0000000000 0.0007803411 0.1940673904
#>
#> $ppv
#> ppv_est ppv_l ppv_u
#> 1.0000000 0.2794604 0.9897390
#>
#> $phi_npv
#> phi lower upper
#> 0.08823529 0.03758016 0.27386671
#>
#> $npv
#> npv_est npv_l npv_u
#> 0.9934025 0.9798027 0.9971794
Note, for walter, no continuity.correction should be used as 0.5 has been used as described by the original paper.
Also comparing to the Zhou’s adjusted methods:
ppv_npv_ci(x1 = 31, n1 = 34, x0 = 44, n0 = 44, prevalence = 0.07,
method = "zhou",
continuity.correction = TRUE)
#> $method
#> [1] "zhou"
#>
#> $sensitivity
#> [1] 0.8699646
#>
#> $specificity
#> [1] 0.9598522
#>
#> $ppv
#> ppv_est ppv_l ppv_u
#> 0.6199169 0.2921698 0.9476640
#>
#> $npv
#> npv_est npv_l npv_u
#> 0.9899059 0.9816510 0.9981609
#>
#> $ppv_logit_transformed
#> ppv_est ppv_l ppv_u
#> 0.6199169 0.2886800 0.8676335
#>
#> $npv_logit_transformed
#> npv_est npv_l npv_u
#> 0.9899059 0.9772354 0.9955563
Feedback and Report issues
We appreciate any feedback, comments and suggestions. If you have any questions or issues to use the package, please reach out to the developers.