Implementation of the Direct Sampling Spatial Prior.
DSSP
The goal of DSSP is to draw samples from the direct sampling spatial prior model (DSSP) described in White et. al. 2019. The basic model assumes a Gaussian likelihood and derives a spatial prior based on thin-plate splines. Functions are included so that the model can be extended to be used for generalised linear mixed models or Bayesian Hierarchical Models.
Installation
You can install the development version from GitHub with:
# install.packages("devtools")
devtools::install_github("gentrywhite/DSSP")
Example
Short example using the Meuse dataset from the {gstat} package.
data("meuse.all", package = "gstat")
sp::coordinates(meuse.all) <- ~ x + y
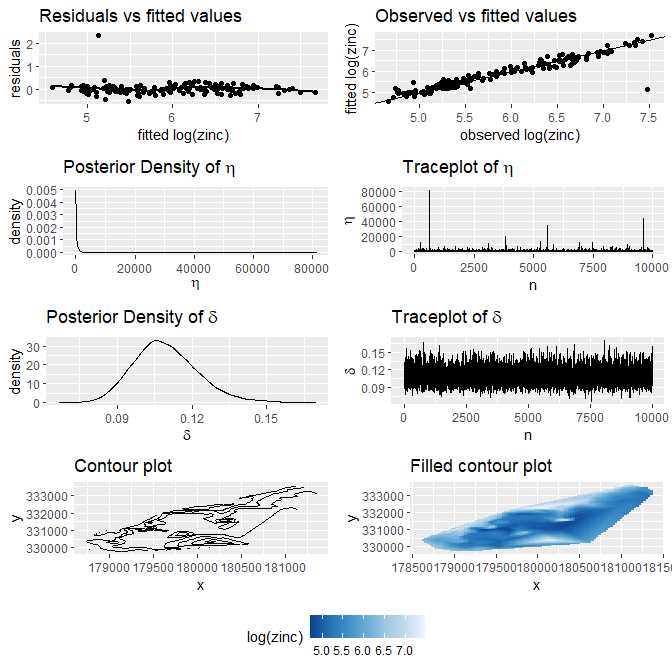
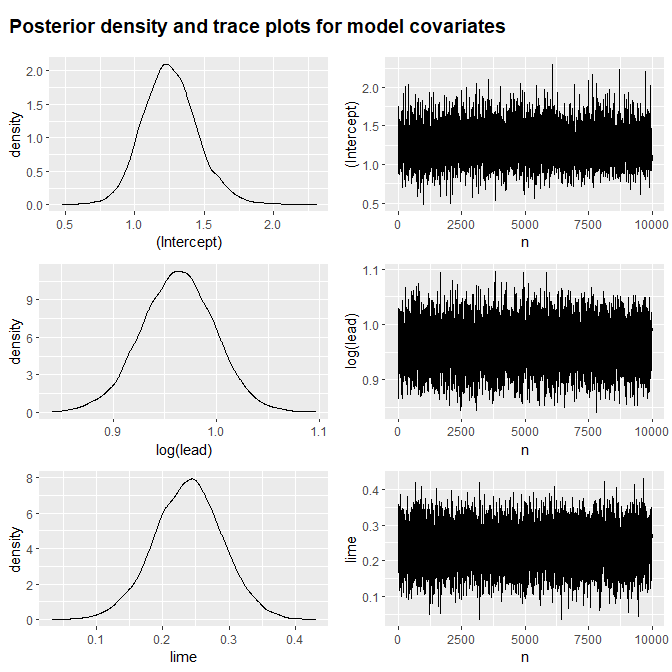
This model does includes two covariates and their posterior densities are summarised in the summary()
library(DSSP)
meuse.fit <- DSSP(
formula = log(zinc) ~ log(lead) + lime, data = meuse.all, N = 10000,
pars = c(0.001, 0.001), log_prior = function(x) -2 * log(1 + x)
)
summary(meuse.fit)
#> Formula: log(zinc) ~ log(lead) + lime
#> Number of observations: 164
#> Number of iterations: 10000
#>
#> Summary of model:
#> Estimate Est.Error l-95% CI u-95% CI ESS
#> eta 334.14 1202.47 3.61 1897.49 10000.00
#> delta 0.11 0.01 0.09 0.14 10000.00
#>
#> Summary of covariates:
#> Estimate Est.Error min q0.025 q0.25 q0.50 q0.75 q0.975 max
#> (Intercept) 1.25 0.20 0.48 0.89 1.12 1.25 1.38 1.67 2.30
#> log(lead) 0.96 0.04 0.84 0.89 0.94 0.96 0.99 1.03 1.10
#> lime 0.24 0.05 0.03 0.14 0.20 0.24 0.27 0.34 0.43
We can inspect several plots for the model using plot().
plot(meuse.fit)
#> `geom_smooth()` using formula 'y ~ x'
Introduction
The Direct Sampling Spatial Prior (DSSP) is based on the thin-plate splines solution to the smoothing problem of minimising the penalised sum of squares
which can be written as
The solution for this problem is
If we assume that the observed data are from a Gaussian distribution
and if we specify the prior for
the resulting posterior of is proportional to
which yields the posterior mean with the same solution as the penalised least squares.
The complete model is specified with a Gaussian likelihood, the improper prior for , an inverse gaussian prior for
, and a prior for
. With this specification the joint posterior is written
Given this it is possible to derive the set of posterior distributions
which can be sampled directly in sequence to create a draw from the joint posterior
This is the heart of what the function DSSP() does[^1].
[^1]: see G. White, D. Sun, P. Speckman (2019) \<arXiv:1906.05575\> for details.
