Estimation of Environmental Variables and Genetic Parameters.
EstimateBreed 
EstimateBreed is an R package designed to perform analyses and estimate environmental covariates and genetic parameters related to selection strategies and the development of superior genotypes. The package offers two main functionalities:
- Prediction models for environmental covariates and processes.
- Estimation of genetic parameters and selection strategies for developing superior genotypes.
Installation
You can install the development version of EstimateBreed from GitHub with:
if (!requireNamespace("pak", quietly = TRUE)) {
install.packages("pak")
}
pak::pak("willyanjnr/EstimateBreed")
Genotype selection
Obtain the genetic selection index for resilience (ISGR) for selecting genotypes for environmental stressors, as described by Bandeira et al. (2024).
library(EstimateBreed)
#Obtain environmental deviations
data("desvamb")
DPclim <- with(desvamb,desv_clim(ENV,TMED,PREC))
DPclim
# A tibble: 3 × 5
# ENV STMED TMEDR SPREC PRECIR
# <chr> <dbl> <dbl> <dbl> <dbl>
# 1 E1 2.65 24.8 5.46 339.
# 2 E2 3.65 23.8 5.27 344.
# 3 E3 2.81 24.5 5.47 362.
#Get the ISGR
data("genot")
isgr_index <- with(genot, isgr(GEN,ENV,NG,MG,CICLO))
isgr_index
# Gen Env ISGR
# 26 L454 E1 6.489941
# 22 L455 E1 7.084315
# 19 L541 E1 7.653157
# 18 L367 E1 7.862185
# 16 L380 E1 8.329434
# 12 L393 E1 9.638909
# 10 L439 E1 10.552056
# 28 L298 E3 12.209433
# 30 L358 E2 23.347984
# 29 L346 E2 23.793351
# 27 L195 E2 24.719927
# 25 L179 E2 25.747317
# 24 L359 E2 26.300686
# 23 L345 E2 26.886419
# 1 L445 E1 27.255375
# 21 L185 E2 28.211433
# 20 L310 E2 28.942165
# 17 L178 E2 31.418785
# 15 L261 E2 33.424611
# 14 L269 E2 34.605133
# 13 L209 E2 35.959423
# 11 L263 E2 39.127798
# 9 L201 E2 43.145922
# 8 L299 E2 45.686042
# 7 L152 E2 48.926278
# 6 L26 E2 52.988109
# 5 L166 E2 57.596139
# 4 L155 E2 64.251152
# 3 L277 E2 74.756384
# 2 L162 E2 86.543916
Selection of transgressive genotypes with the selection differential (mean and standard deviations).
library(EstimateBreed)
Gen <- paste0("G", 1:20)
Var <- round(rnorm(20, mean = 3.5, sd = 0.8), 2)
Control <- rep(3.8, 20)
data <- data.frame(Gen,Var,Control)
with(data,transg(Gen,Var,Control))
Returns the general parameters and the genotypes selected for each treshold. Also plot a representative graph of the selected genotypes based on the mean and standard deviations.
---------------------------------------------------------------------
Selection of Transgressive Genotypes - Selection Differential (SD)
---------------------------------------------------------------------
Parameters:
---------------------------------------------------------------------
Overall Mean : 3.566
Control Mean : 3.800
Standard Deviation : 0.603
Mean + 1SD : 4.169
Mean + 2SD : 4.771
Mean + 3SD : 5.374
---------------------------------------------------------------------
Genotypes above each threshold:
---------------------------------------------------------------------
Genotypes above Control Mean : G4, G7, G8, G9, G12, G14,
G20
Genotype above Overall Mean : G4, G7, G8, G9, G12, G14,
G16, G18, G20
Genotypes above Mean + 1SD : G7, G9, G20
Genotypes above Mean + 2SD : G7
Genotypes above Mean + 3SD : None
---------------------------------------------------------------------
Estimation of environmental variables and processes
Predict ∆T to determine the ideal times to apply agricultural pesticides.
library(EstimateBreed)
# Forecasting application conditions
tdelta(-53.696944444444,-28.063888888889,type=1,days=10)
# Retrospective analysis of application conditions
tdelta(-53.6969,-28.0638,type=2,days=10,dates=c("2023-01-01","2023-05-01"))
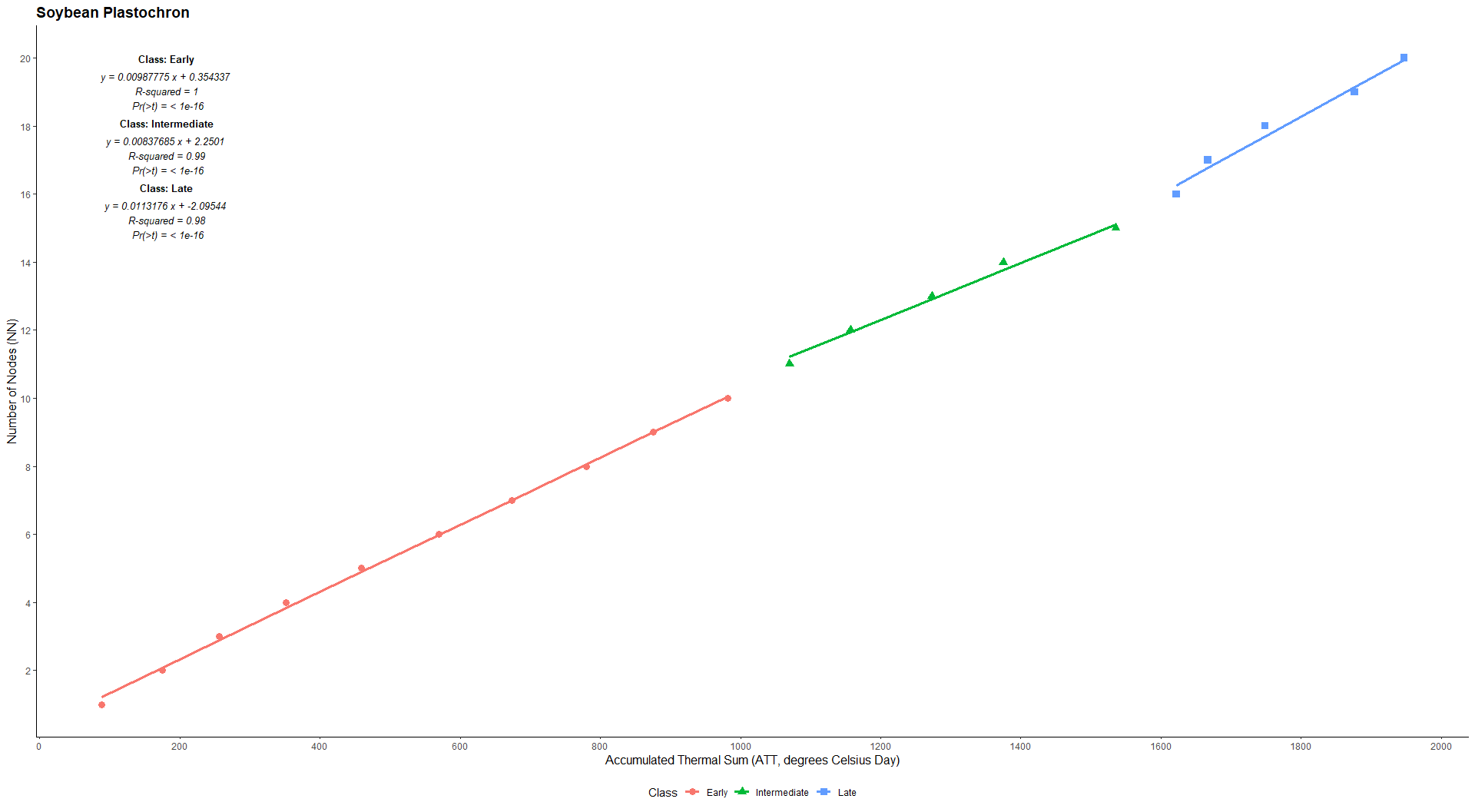
Estimation of soybean plastochron using average air temperature and number of nodes
library(EstimateBreed)
data("pheno")
with(pheno, plast(GEN,TMED,EST,NN,habit="ind",plot=TRUE))
#
Documentation
Complete documentation can be found when using the package within R.
Citing
When citing this package, please use,
library(EstimateBreed)
citation("EstimateBreed")
To cite package ‘EstimateBreed’ in publications use:
Willyan Jr. A. Bandeira, Ivan R. Carvalho, Murilo V. Loro, Leonardo
C. Pradebon, José A. G. da Silva (2025). _EstimateBreed: Estimation
of Environmental Variables and Genetic Parameters_. R package
version 0.1.0, <https://github.com/willyanjnr/EstimateBreed>.
A BibTeX entry for LaTeX users is
@Manual{,
title = {EstimateBreed: Estimation of Environmental Variables and Genetic Parameters},
author = {{Willyan Jr. A. Bandeira} and {Ivan R. Carvalho} and {Murilo V. Loro} and {Leonardo C. Pradebon} and {José A. G. da Silva}},
year = {2025},
note = {R package version 0.1.0},
url = {https://github.com/willyanjnr/EstimateBreed},
}
Getting Help
- If you find any errors, please make a report with the commands used so that we can repeat, check and adjust the functions! Send it to github or send an email to [email protected].