Fundamental Clustering Problems Suite.
FCPS
Fundamental Clustering Problems Suite
The package provides over sixty state-of-the-art clustering algorithms for unsupervised machine learning published in .
Table of contents
Description
The Fundamental Clustering Problems Suite (FCPS) summaries over sixty state-of-the-art clustering algorithms available in R language. An important advantage is that the input and output of clustering algorithms is simplified and consistent in order to enable users a swift execution of cluster analysis. By combining mirrored-density plots (MD plots) with statistical testing FCPS provides a tool to investigate the cluster tendency quickly prior to the cluster analysis itself . Common clustering challenges can be generated with arbitrary sample size
. Additionally, FCPS sums 26 indicators with the goal to estimate the number of clusters up and provides an appropriate implementation of the clustering accuracy for more than two clusters
. A subset of methods was used in a benchmarking of algorithms published in
.
Installation
Installation using CRAN
Install automatically with all dependencies via
install.packages("FCPS",dependencies = T)
# Optionally, for the automatic installation
# of all suggested packages:
Suggested=c("kernlab", "cclust", "dbscan", "kohonen",
"MCL", "ADPclust", "cluster", "DatabionicSwarm",
"orclus", "subspace", "flexclust", "ABCanalysis",
"apcluster", "pracma", "EMCluster", "pdfCluster", "parallelDist",
"plotly", "ProjectionBasedClustering", "GeneralizedUmatrix",
"mstknnclust", "densityClust", "parallel", "energy", "R.utils",
"tclust", "Spectrum", "genie", "protoclust", "fastcluster",
"clusterability", "signal", "reshape2", "PPCI", "clustrd", "smacof",
"rgl", "prclust", "dendextend",
"moments", "prabclus", "VarSelLCM", "sparcl", "mixtools",
"HDclassif", "clustvarsel", "knitr", "rmarkdown")
for(i in 1:length(Suggested)) {
if (!requireNamespace(Suggested[i], quietly = TRUE)) {
message(paste("Installing the package", Suggested[i]))
install.packages(Suggested[i], dependencies = T)
}
}
Installation using Github
Please note, that dependecies have to be installed manually.
remotes::install_github("Mthrun/FCPS")
Installation using R Studio
Please note, that dependecies have to be installed manually.
Tools -> Install Packages -> Repository (CRAN) -> FCPS
Tutorial Examples
The tutorial with several examples can be found on in the vignette on CRAN:
https://cran.r-project.org/web/packages/FCPS/vignettes/FCPS.html
Manual
The full manual for users or developers is available here: https://cran.r-project.org/web/packages/FCPS/FCPS.pdf
Use Cases
Cluster Analysis of High-dimensional Data
The package FCPS provides a clear and consistent access to state-of-the-art clustering algorithms:
library(FCPS)
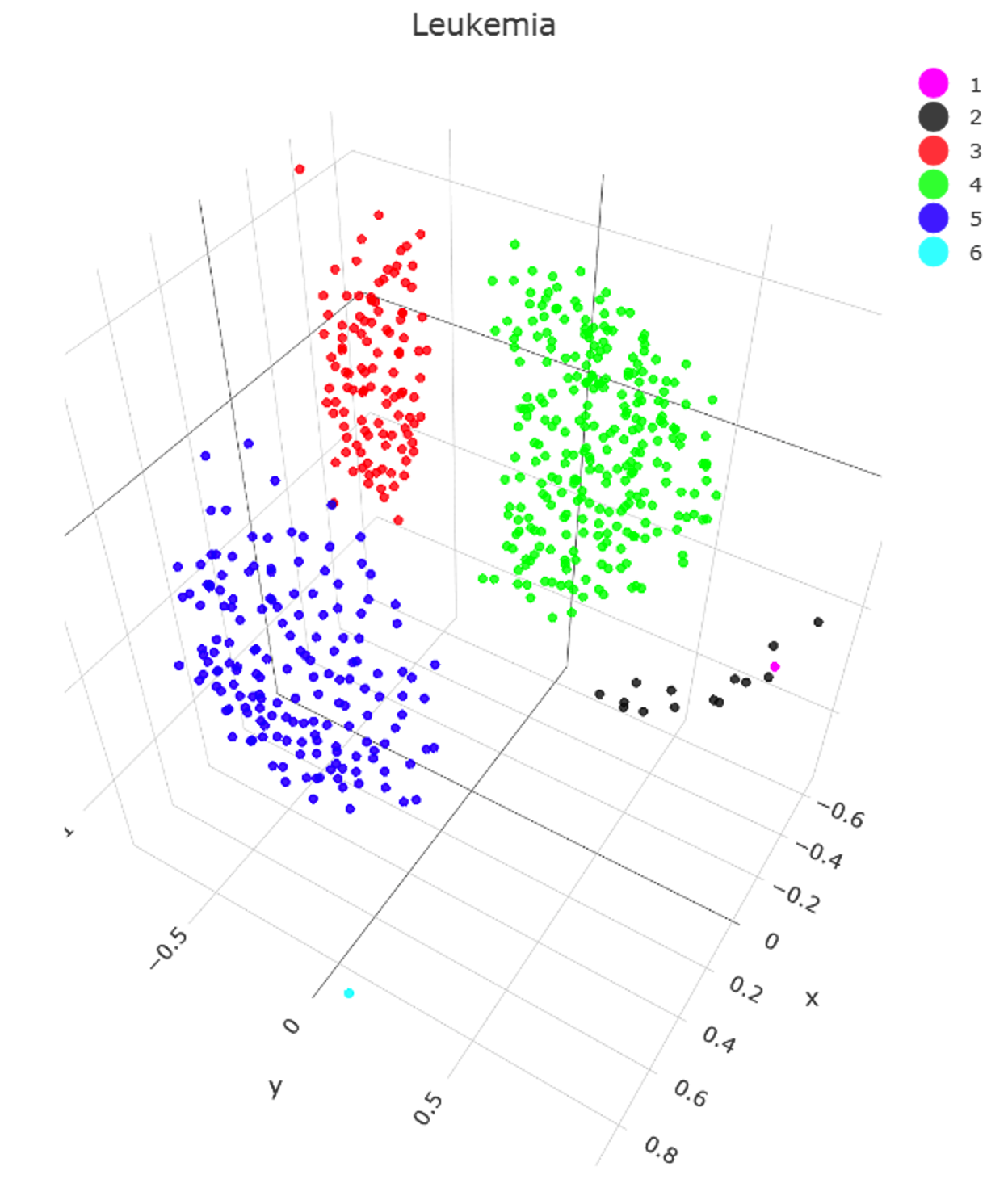
data("Leukemia")
Data=Leukemia$Distance
Classification=Leukemia$Cls
ClusterNo=6
CA=ADPclustering(Leukemia$DistanceMatrix,ClusterNo)
Cls=ClusterRenameDescendingSize(CA$Cls)
ClusterPlotMDS(Data,Cls,main = ’Leukemia’,Plotter3D = ’plotly’)
ClusterAccuracy(Cls,Classification)
[1] 0.9963899
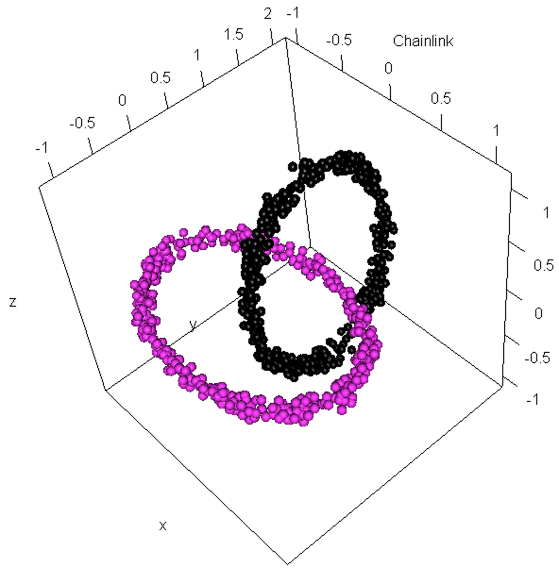
Generating Typical Challenges for Clustering Algorithms
Several clustering challenge can be generated with an arbitrary sample size:
set.seed(600)
library(FCPS)
DataList=ClusterChallenge("Chainlink", SampleSize = 750,
PlotIt=TRUE)
Data=DataList$Chainlink
Cls=DataList$Cls
> ClusterCount(Cls)
$CountPerCluster
$NumberOfClusters
$ClusterPercentages
[1] 377 373
[1] 2
[1] 50.26667 49.73333
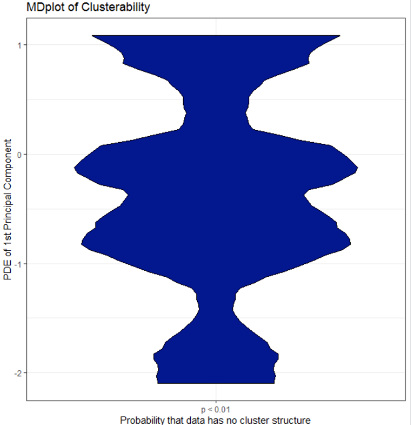
Cluster-Tendency
For many applications, it is crucial to decide if a dataset possesses cluster structures:
library(FCPS)
set.seed(600)
DataList=ClusterChallenge("Chainlink",SampleSize = 750)
Data=DataList$Chainlink
Cls=DataList$Cls
library(ggplot2)
ClusterabilityMDplot(Data)+theme_bw()
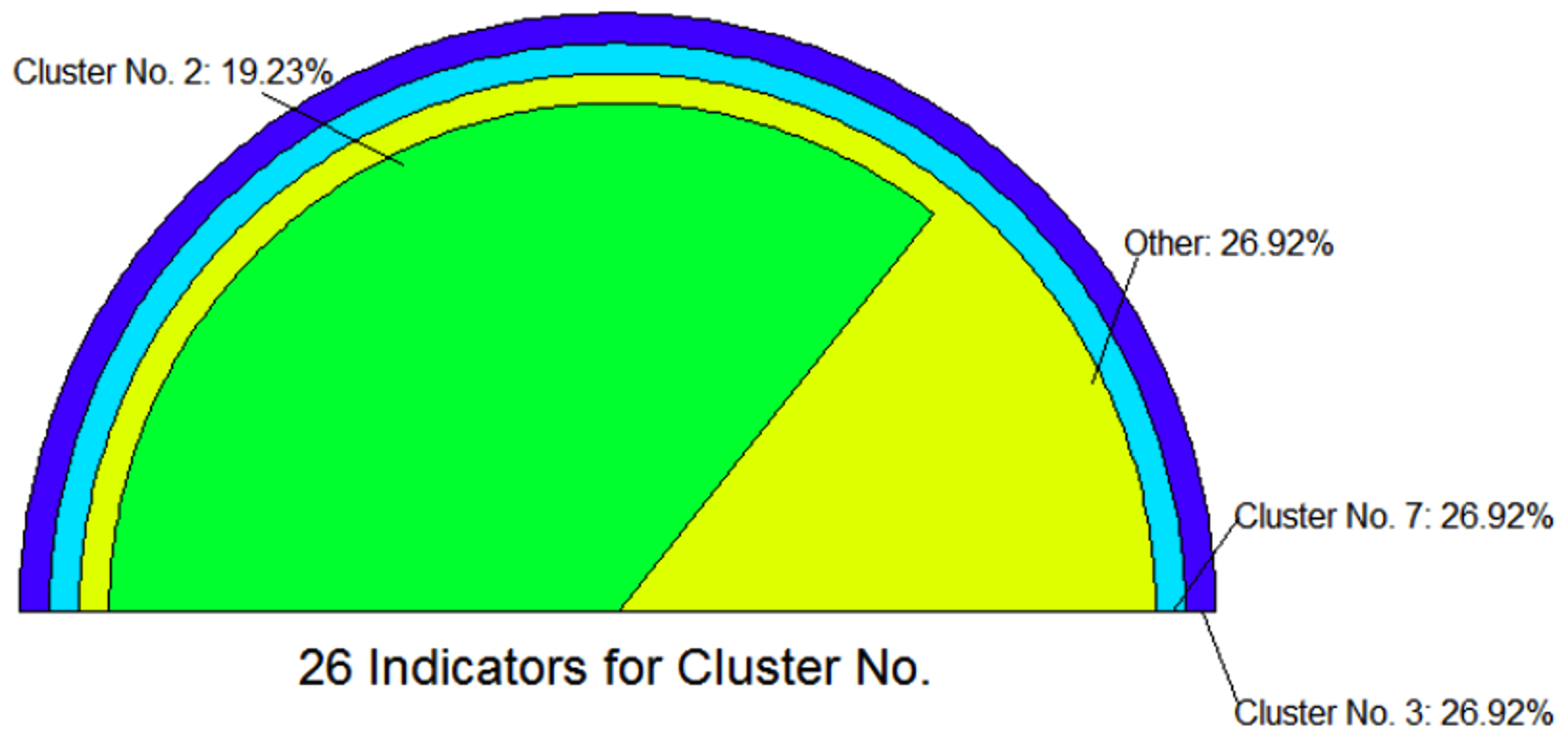
Estimation of Number of Clusters
The “FCPS” package provides up to 26 indicators to determine the number of clusters:
library(FCPS)
set.seed(135)
DataList=ClusterChallenge("Chainlink",SampleSize = 900)
Data=DataList$Chainlink
Cls=DataList$Cls
Tree=HierarchicalClustering(Data,0,"SingleL")[[3]]
ClusterDendrogram(Tree,4,main="Single Linkage")
MaximumNumber=7
clsm <- matrix(data = 0, nrow = dim(Data)[1], ncol = MaximumNumber)
for (i in 2:(MaximumNumber+1)) {
clsm[,i-1] <- cutree(Tree,i)
}
out=ClusterNoEstimation(Data, ClsMatrix = clsm,
MaxClusterNo = MaximumNumber, PlotIt = TRUE)
Additional information
| Authors website | http://www.deepbionics.org/ |
|---|---|
| License | GPL-3 |
| Dependencies | R (>= 3.5.0) |
| Bug reports | https://github.com/Mthrun/FCPS/issues |
References
- [Thrun/Stier, 2021] Thrun, M. C., & Stier, Q.: Fundamental Clustering Algorithms Suite SoftwareX, Vol. 13(C), pp. 100642. doi 10.1016/j.softx.2020.100642, 2021.
- [Thrun, 2020] Thrun, M. C.: Improving the Sensitivity of Statistical Testing for Clusterability with Mirrored-Density Plot, in Archambault, D., Nabney, I. & Peltonen, J. (eds.), Machine Learning Methods in Visualisation for Big Data, DOI 10.2312/mlvis.20201102, The Eurographics Association, Norrköping , Sweden, May, 2020.
- [Thrun/Ultsch, 2020a] Thrun, M. C., & Ultsch, A.: Clustering Benchmark Datasets Exploiting the Fundamental Clustering Problems, Data in Brief,Vol. 30(C), pp. 105501, DOI 10.1016/j.dib.2020.105501 , 2020.
- [Thrun/Ultsch, 2021] Thrun, M. C., and Ultsch, A.: Swarm Intelligence for Self-Organized Clustering, Artificial Intelligence, Vol. 290, pp. 103237, \doi{10.1016/j.artint.2020.103237}, 2021.
- [Thrun/Ultsch, 2020b] Thrun, M. C., & Ultsch, A. : Using Projection based Clustering to Find Distance and Density based Clusters in High-Dimensional Data, Journal of Classification, \doi{10.1007/s00357-020-09373-2}, Springer, 2020.