Simple Peak Alignment for Gas-Chromatography Data.
GCalignR 
GCalignR provides simple functions to align peak lists obtained from Gas Chromatography Flame Ionization Detectors (GC-FID) based on retention times and plots to evaluate the quality of the alignment. The package supports any other one-dimensional chromatography technique that enables the user to create a peak list with at least one column specifying retention times as illustrated below.
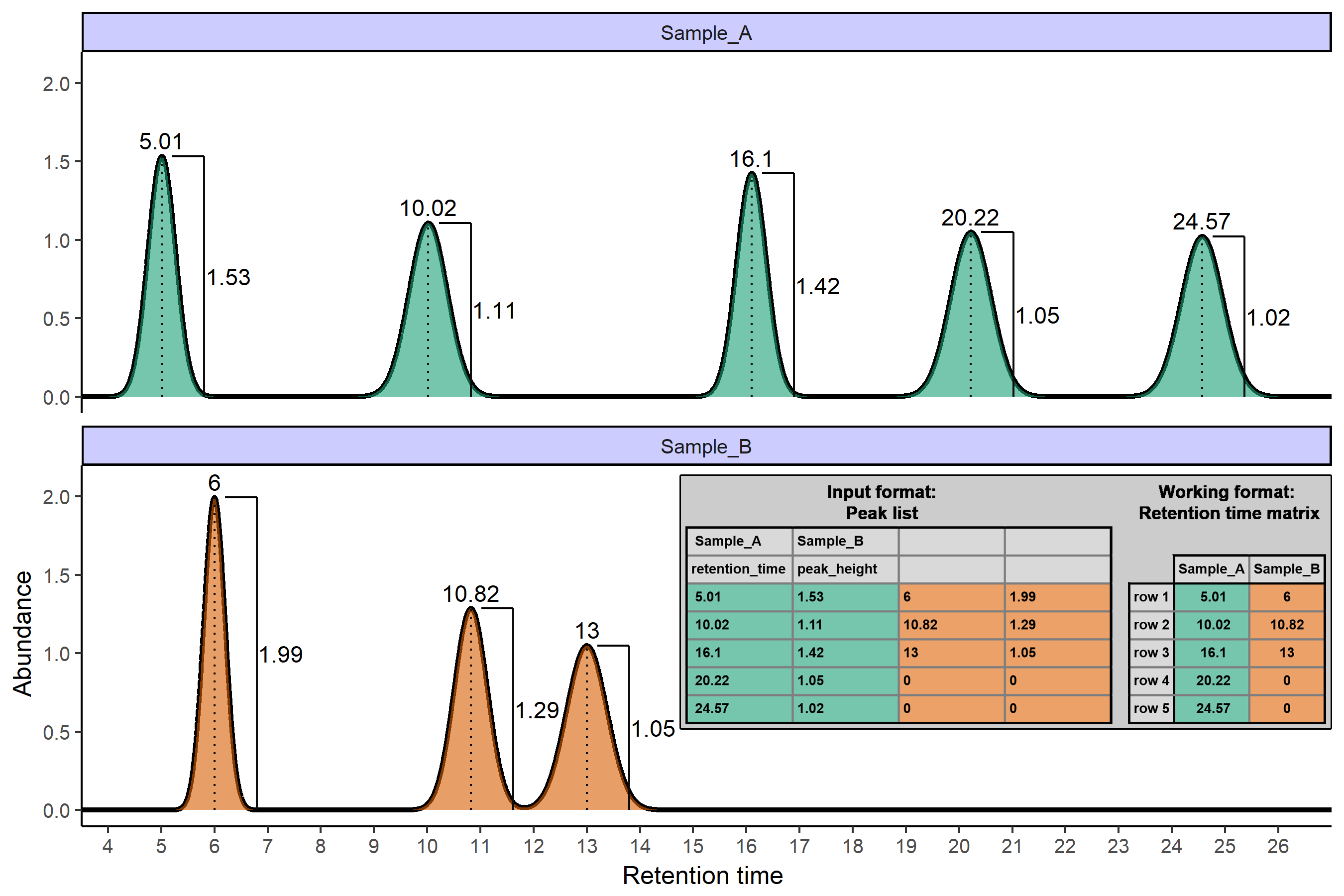
As with other software you need to get used to the input format which is shown in the illustration:
- Row 1: Sample names
- Row 2: Variable names
- Row 3-N: GC data
- Each block belongs to a sample as shown for sample A (green) and sample B (orange) above
Installing GCalignR:
The latest release v1.0.6 is on CRAN. Click here for an overview of past releases and a brief description of applied changes.
install.packages("GCalignR", dependencies = T)
The current developmental version is identical to the CRAN release
if (!("devtools" %in% rownames(installed.packages()))) {
install.packages("devtools")
} else if (packageVersion("devtools") < 1.6) {
install.packages("devtools")
}
devtools::install_github("mottensmann/GCalignR", build_vignettes = TRUE)
Get started with GCalignR
To get started read the vignettes:
browseVignettes("GCalignR")
Basic usage of the main function to align peaks:
data: Path to a text file (see input format above), or list of data frames, each corresponding to a samplert_col_name: column name of retention time valuesmax_linear_shift: Here, no adjustment of systematic linear driftmax_diff_peak2mean: Here, sort all peaks strictly by retention timemin_diff_peak2peak: Here, try to merge peaks when rt differs by less than 0.1
library(GCalignR)
#> Warning: package 'GCalignR' was built under R version 4.4.1
aligned <- align_chromatograms(data = peak_data[1:4], # list of data frame
rt_col_name = "time", # retention time
max_linear_shift = 0, #
max_diff_peak2mean = 0,
min_diff_peak2peak = 0.08)
#> Run GCalignR
#> Start: 2024-07-03 14:53:38
#>
#> Data for 4 samples loaded.
#> No reference was specified. Hence, a reference will be selected automatically ...
#>
#> 'C2' was selected on the basis of highest average similarity to all samples (score = 0.06).
#> Start correcting linear shifts with "C2" as a reference ...
#>
#> Start aligning peaks ... this might take a while!
#>
#> Merge redundant rows ...
#>
#> Alignment completed!
#> Time: 2024-07-03 14:53:41
The parameter values above differ from the defaults shown in the paper and the package vignette. In a nutshell, we now suggest in most cases to set max_diff_peak2mean = 0. This way peaks are first simply sorted based on the given retention time value and then purely min_diff_peak2peak specifies which peaks will be evaluated for a merge. Additionally, this enables the possibility for a considerable boost in computation speed of the first alignment steps (available since version 1.0.5, currently only on GitHub!)
If you encounter bugs or if you have any suggestions for improvement (for instance on how to speed up the algorithm!), just contact meinolf.ottensmann[at]web.de
Also I´m happy to provide help if you can´t get it to work. Usually it is easy to solve small problems. However, in order to simplify this process please send a short description of the problem along with the code you have been using as a script file (.R) together with a minimal example input file (.txt).
Published paper
#> Warning: package 'ggplot2' was built under R version 4.4.1

