'GeneSelectR' - Comprehensive Feature Selection Workflow for Bulk RNAseq Datasets.
GeneSelectR
Overview

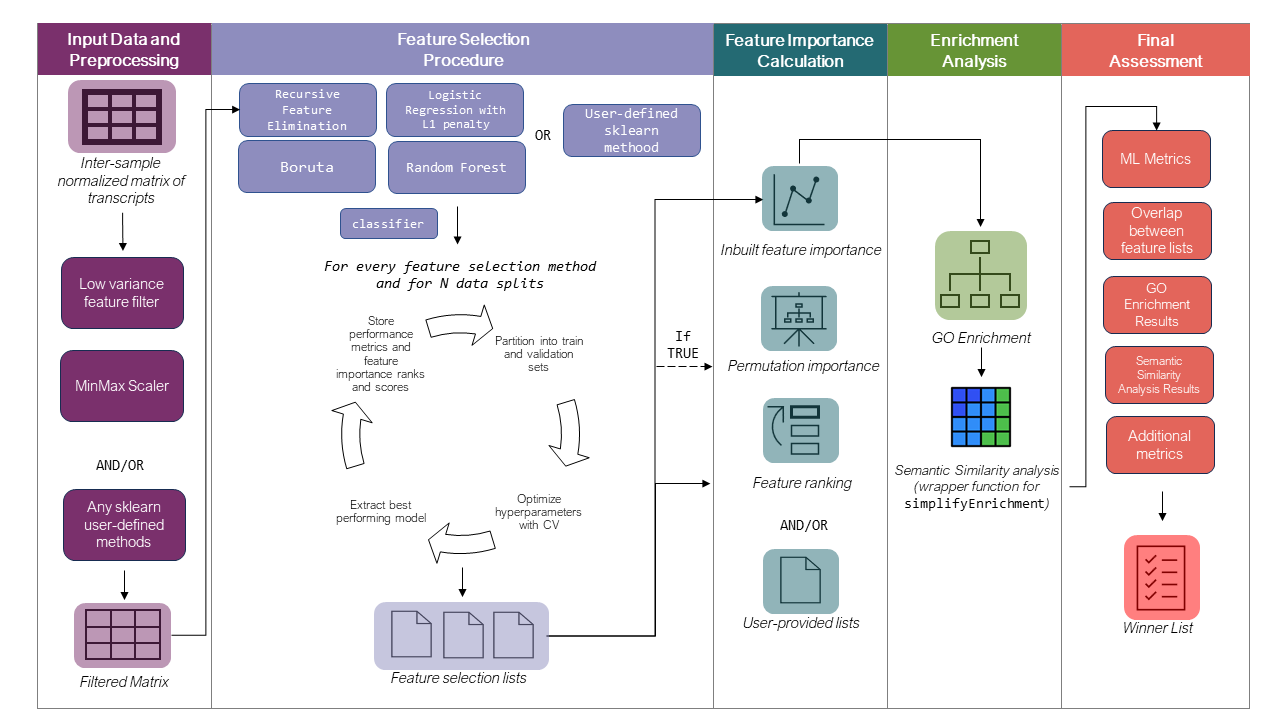
GeneSelectR is an R package designed to streamline the process of gene selection and evaluation in bulk RNAseq datasets. Built on top of the powerful scikit-learn Python library via the reticulate package, GeneSelectR offers a seamless integration of machine learning and bioinformatics capabilities in a single workflow.
Features
Comprehensive Workflow GeneSelectR provides an end-to-end solution for feature selection, combining the machine learning prowess of scikit-learn with the bioinformatics utilities of R packages like clusterprofiler and simplifyEnrichment.
Customizable Yet User-Friendly
While GeneSelectR offers a high degree of customization to cater to specific research needs, it also comes with preset configurations that are suitable for most use-cases, making it accessible for both novice and experienced users.
Diverse Feature Selection Methods
The package includes a variety of inbuilt feature selection methods, such as:
- SelectFromModel with RandomForest
- SelectFromModel with Logistic Regression (L1 penalty)
- Boruta
- Univariate Filtering
Main Functionality
The core function, GeneSelectR, performs gene selection using various methods and evaluates their performance through cross-validation. It also supports hyperparameter tuning, permutation feature importance calculation, and more.
Installation
GeneSelectR depends on reticulate that creates a conda working environment. Please, install Anaconda distribution before you proceed. You can install the development version of GeneSelectR from GitHub with:
# install.packages("devtools")
devtools::install_github("dzhakparov/GeneSelectR")
Usage and Example
A tutorial detailing how to use GeneSelectR can be accessed in this vignette.
Docker Image
GeneSelectR is available as a container image on Docker Hub. You can pull the image using the following command:
docker pull dzhakparov/geneselectr-image:latest
docker run -e PASSWORD=your_password -p 8787:8787 dzhakparov/geneselectr-image:latest
After running these commands, open your browser and go to localhost:8787 (http//local-ip-address:8787 in Windows). You will be prompted to enter username and password. The default username is rstudio and the password is the one you specified in the command above.
Citation
Please cite the following paper if you use GeneSelectR in your research:
Feedback and Contribution
Any feedback is welcome and appreciated! Feel free to create issues or pull requests. For any other questions please write to: [email protected].