Linear Approach to Threshold with Ergodic Rate for Reaction Times.
LATERmodel
The LATERmodel R package is an open-source implementation of Roger Carpenter’s Linear Approach to Threshold with Ergodic Rate (LATER) model (R. Carpenter (1981), Noorani and Carpenter (2016)). This package enables the easy visualisation of reaction time data in LATER’s signature reciprobit space, as well as estimating parameters to fit the model to datasets, comparing raw datasets, comparing fits (i.e., is the dataset better explained by a shift or a swivel?), adding an early component, etc.
This package also includes two canonical datasets digitised from R. H. S. Carpenter and Williams (1995) and Reddi, Asrress, and Carpenter (2003).
Installation
You can install LATERmodel with:
install.packages("LATERmodel")
Example
Load digitised data from Figure 1 in Carpenter and Williams (1995):
library(LATERmodel)
data(carpenter_williams_1995)
Extract data corresponding only to participant a (Figure 1.a):
raw_data <- subset(carpenter_williams_1995, participant == "a")
The data analysis functions within this package require the raw data to first undergo pre-processing using the prepare_data function. We pass our raw_data variable as the argument to the raw_data parameter of prepare_data to perform such pre-processing:
data <- prepare_data(raw_data = raw_data)
Fit each condition individually (no shared parameters between them), and include an early component for all of them:
data_fit <- individual_later_fit(data, with_early_component = TRUE)
Visualise the raw data and the best individual fits:
reciprobit_plot(data, data_fit)
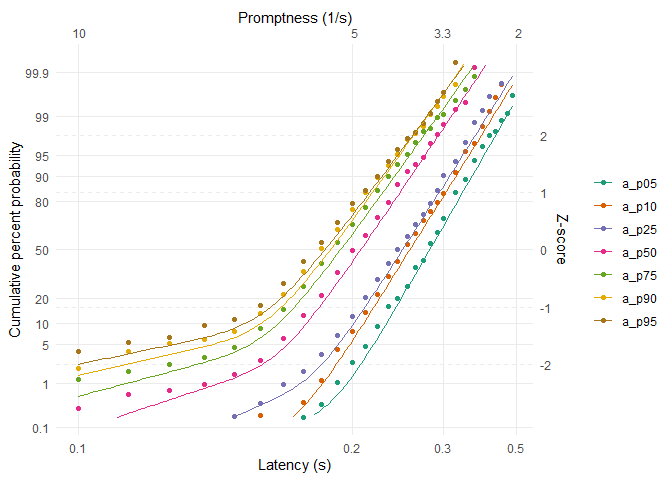
For a more detailed analysis using the LATERmodel R package, see this article.
Carpenter, R. H. S., and M. L. L. Williams. 1995. “Neural Computation of Log Likelihood in Control of Saccadic Eye Movements.” Nature 377 (6544): 59–62. https://doi.org/10.1038/377059a0.
Carpenter, RHS. 1981. “Eye Movements: Cognition and Visual Perception.” Oculomotor Procrastination (Fisher DF, Monty RA, Senders JW, Eds) Pp 237: 246.
Noorani, Imran, and R. H. S. Carpenter. 2016. “The LATER Model of Reaction Time and Decision.” Neuroscience & Biobehavioral Reviews 64 (May): 229–51. https://doi.org/10.1016/j.neubiorev.2016.02.018.
Reddi, B. a. J., K. N. Asrress, and R. H. S. Carpenter. 2003. “Accuracy, Information, and Response Time in a Saccadic Decision Task.” Journal of Neurophysiology 90 (5): 3538–46. https://doi.org/10.1152/jn.00689.2002.