Modeling for Single-Cell Open Chromatin Analysis.
MOCHA: Model-based single cell Open CHromatin Analysis
Table of Contents
- Introduction
- Installation
- Usage: Package Vignette on COVID PASC dataset
- Tips: Result formats
- Contact
- License
Introduction
MOCHA is an R package containing a novel single-cell peak-calling algorithm that leverages single-cell information to determine whether a particular genomic region is open by calculating two measures of intensities, and using these to call peaks via a hierarchical model.
Install package and load library
You can install MOCHA directly from CRAN using the following command.
install.packages('MOCHA')
Usage
Please view the example usage found in the vignette found under vignettes/COVID-walkthrough.html.
The example usage demonstrates this workflow:
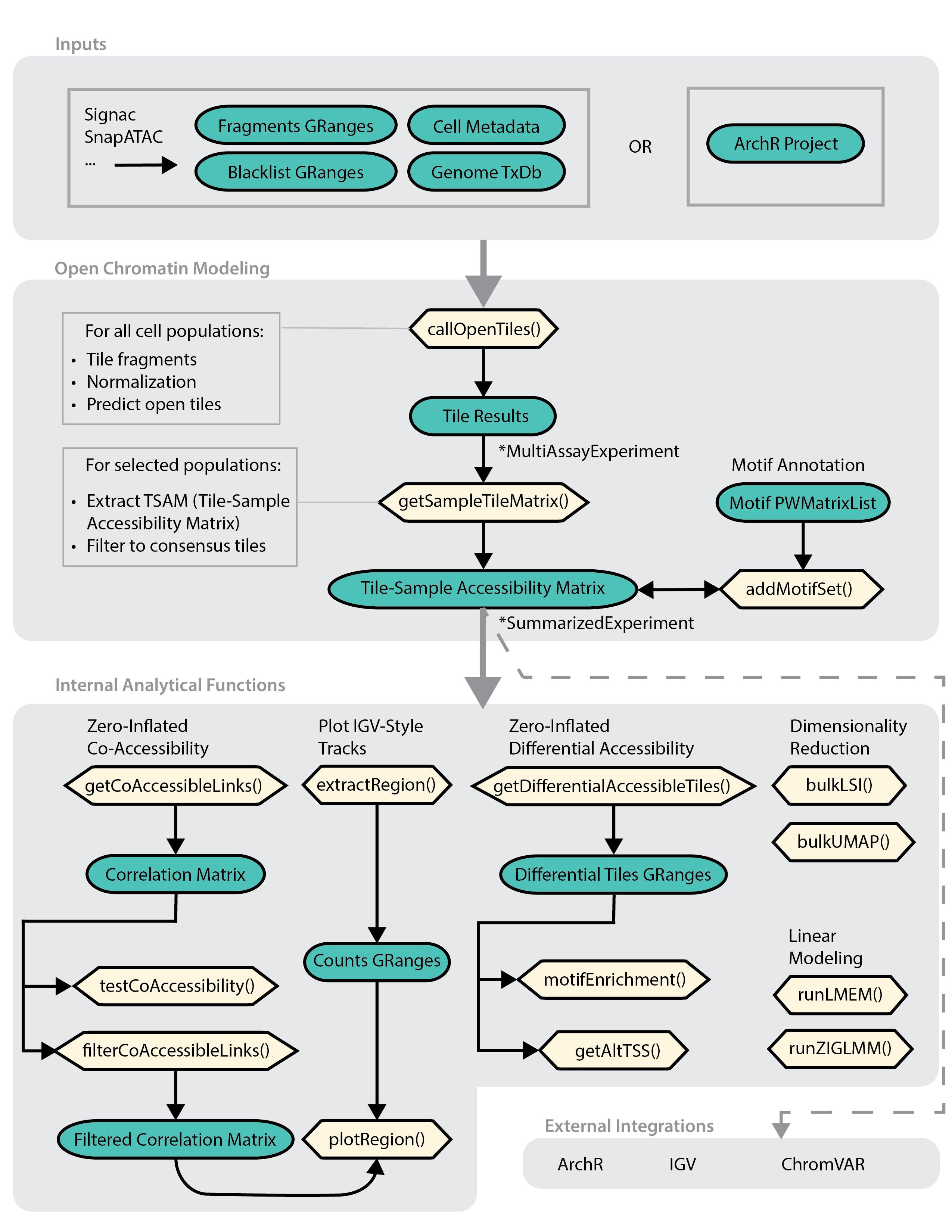
Tips: Result formats
While the pipeline can be run function-to-function, you may wish to inspect intermediate results or use end results for your own custom analyses. All MOCHA outputs use common Bioconductor data structures. We also provide some getters for accessing specific results.
callOpenTiles results
The output of the first step of the pipeline, MOCHA::callOpenTiles is a MultiAssayExperiment organizing your open-tiles by cell population. This is the input to the next function, MOCHA::getSampleTileMatrices.
Sample-level metadata can be accessed using colData(tileResults).
A specific cell population's results are stored as a RaggedExperiment.
This object stores ranged data alongside the sample-level metadata.
getSampleTileMatrix results
The output of getSampleTileMatrix is a SummarizedExperiment organizing the TSAM (Tile Sample Accessibility Matrix) by cell population. This is the input to MOCHA::getDifferentialAccessibleTiles and other downstream analyses.
## Subset results for Chr 4
> SampleTileMatrices[grep('chr4', rownames(SampleTileMatrices)),]
class: RangedSummarizedExperiment
dim: 8112 39
metadata(6): CellCounts FragmentCounts ... OrgDb Directory
assays(1): CD16 Mono
rownames(8112): chr4:10005500-10005999 chr4:10006000-10006499 ...
chr4:99961000-99961499 chr4:99961500-99961999
rowData names(1): CD16 Mono
colnames(39): B011-AP0C1W3 B011-AP0C1W8 ... FSQEAZ0C2D3-02
FSQFAZ0BZJQ-02
colData names(186): Sample well_id ... ATAC_WellID AIFI.Batch
It also holds metadata related to the genome, transcript database, and annotations: metadata(SampleTileMatrices)
getDifferentialAccessibleTiles results
Results of MOCHA::getDifferentialAccessibleTiles is given either as a data.table or a 'granges' and can be filtered accordingly:
> head(plyranges::filter(differentials, seqnames =='chr4' & FDR < 0.2))
GRanges object with 6 ranges and 13 metadata columns:
seqnames ranges strand | Tile
<Rle> <IRanges> <Rle> | <character>
[1] chr4 10023500-10023999 * | chr4:10023500-10023999
[2] chr4 10024000-10024499 * | chr4:10024000-10024499
[3] chr4 10063500-10063999 * | chr4:10063500-10063999
[4] chr4 10096500-10096999 * | chr4:10096500-10096999
[5] chr4 1010500-1010999 * | chr4:1010500-1010999
[6] chr4 101346000-101346499 * | chr4:101346000-10134..
CellPopulation Foreground Background P_value Test_Statistic
<character> <character> <character> <numeric> <numeric>
[1] CD16 Mono Positive Negative 0.002610899 11.89612
[2] CD16 Mono Positive Negative 0.023560228 7.49639
[3] CD16 Mono Positive Negative 0.000421733 12.43336
[4] CD16 Mono Positive Negative 0.006561717 10.05301
[5] CD16 Mono Positive Negative 0.016701496 5.72747
[6] CD16 Mono Positive Negative 0.006274534 7.46972
FDR Log2FC_C MeanDiff Avg_Intensity_Case Pct0_Case
<numeric> <numeric> <numeric> <numeric> <numeric>
[1] 0.0788621 1.023296 2.9715709 12.3478 0.0000000
[2] 0.1910923 0.648633 -0.0879542 12.2027 0.0588235
[3] 0.0451423 0.786501 0.7773734 12.8422 0.0000000
[4] 0.1124369 0.739042 1.3575321 13.0183 0.0000000
[5] 0.1628641 -0.476150 -0.4293368 13.6077 0.0000000
[6] 0.1091473 -0.397274 -0.3913687 13.1689 0.0000000
Avg_Intensity_Control Pct0_Control
<numeric> <numeric>
[1] 11.1988 0.1818182
[2] 11.4155 0.0000000
[3] 12.0881 0.0000000
[4] 12.2886 0.0454545
[5] 14.2039 0.0000000
[6] 13.5651 0.0000000
-------
seqinfo: 24 sequences from an unspecified genome; no seqlengths
getCoAccessibleLinks results
Results of MOCHA::getCoAccessibleLinks is given as a data.frame and can be filtered further according to correlation using MOCHA::filterCoAccessibleLinks.
> head(links)
Correlation Tile1 Tile2
1: 0.06203363 chr4:10023500-10023999 chr4:10005500-10005999
2: -0.07812306 chr4:10023500-10023999 chr4:10006000-10006499
3: 0.06192133 chr4:10023500-10023999 chr4:10006500-10006999
4: 0.25329153 chr4:10023500-10023999 chr4:10007000-10007499
5: 0.09938466 chr4:10023500-10023999 chr4:10013500-10013999
6: 0.03643015 chr4:10023500-10023999 chr4:10015000-10015499
> MOCHA::filterCoAccessibleLinks(links, threshold = 0.4)
Correlation Tile1 Tile2 chr start end
1: 0.5035455 chr4:10023500-10023999 chr4:10024000-10024499 chr4 10023500 10024499
2: 0.5121115 chr4:10023500-10023999 chr4:10063500-10063999 chr4 10023500 10063999
3: 0.4059254 chr4:10023500-10023999 chr4:10078000-10078499 chr4 10023500 10078499
4: 0.4003500 chr4:10023500-10023999 chr4:10096500-10096999 chr4 10023500 10096999
5: 0.4367843 chr4:10023500-10023999 chr4:10106500-10106999 chr4 10023500 10106999
Contact
To contact the developers on issues and feature requests, please contact us via the discussions tab for feature requests, or open issues for any bugs.
License
MOCHA follows the Allen Institute Software License - full information about the license can be found on the LICENSE file.