Observers Needed to Evaluate Subjective Tests.
ONEST
The Observers Needed to Evaluate Subjective Tests software implements a statistical method in Reisenbichler et al. (2020[1]), to determine the minimum number of evaluators needed to estimate agreement involving a large number of raters. This method could be utilized by regulatory agencies, such as the FDA, when evaluating agreement levels of a newly proposed subjective laboratory test. Input to the program should be binary(1/0) pathology data, where “0” may stand for negative and “1” for positive. The example datasets in this software are from Rimm et al. (2017[2]) (the SP142 assay), and Reisenbichler et al. 2020. This program can run in R version 3.5.0 and above.
Installation
You can install RPEXE.RPEXT from github with:
# install.packages("devtools")
devtools::install_github("hangangtrue/ONEST")
You can also install the released version of ONEST from CRAN with:
install.packages("ONEST")
Example
This is a basic example which shows you how this software works:
library(ONEST)
# load in the sp142_bin dataset
data('sp142_bin')
ONEST_main(sp142_bin)
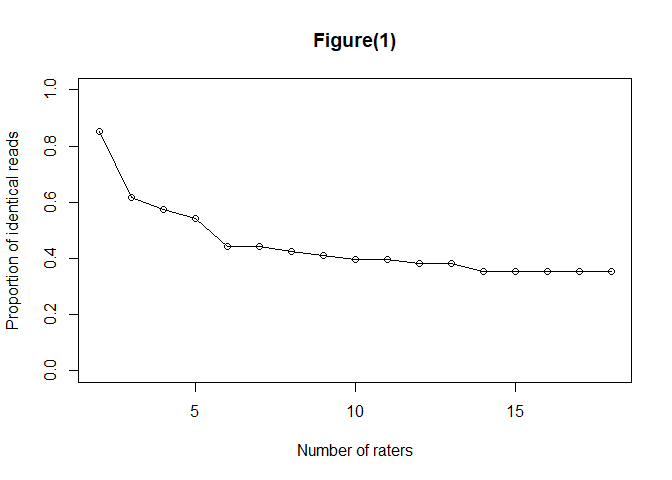
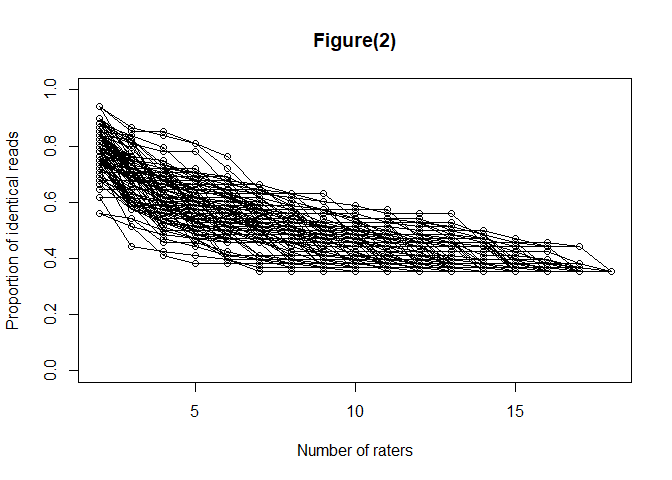
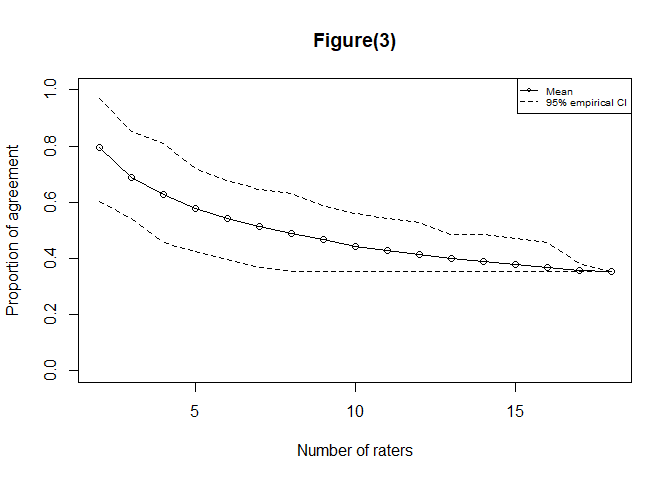
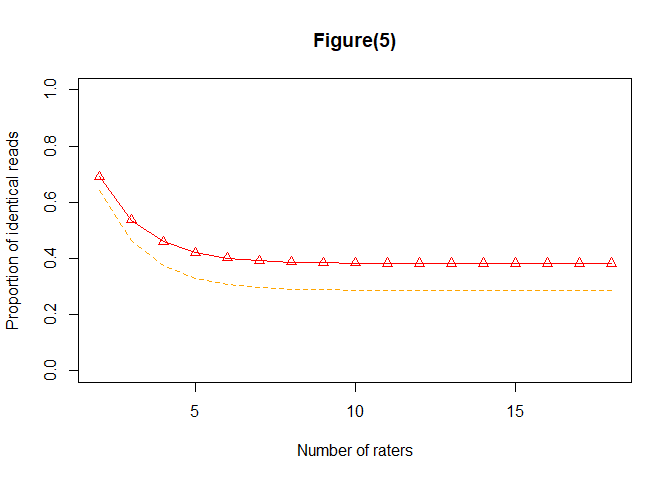
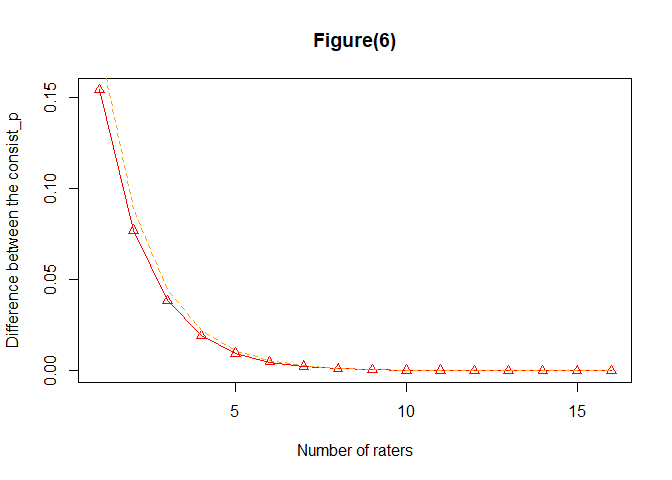
#> $consistency
#> consist_p consist_low
#> [1,] 0.6911795 0.6427088
#> [2,] 0.5367693 0.4640632
#> [3,] 0.4595634 0.3747395
#> [4,] 0.4209597 0.3300768
#> [5,] 0.4016573 0.3077448
#> [6,] 0.3920057 0.2965783
#> [7,] 0.3871797 0.2909948
#> [8,] 0.3847665 0.2882029
#> [9,] 0.3835598 0.2868068
#> [10,] 0.3829564 0.2861087
#> [11,] 0.3826547 0.2857597
#> [12,] 0.3825039 0.2855851
#> [13,] 0.3824284 0.2854978
#> [14,] 0.3823907 0.2854542
#> [15,] 0.3823718 0.2854324
#> [16,] 0.3823624 0.2854214
#> [17,] 0.3823577 0.2854160
#>
#> $difference
#> diff_consist diff_high
#> [1,] -1.544102e-01 1.786456e-01
#> [2,] -7.720588e-02 8.932368e-02
#> [3,] -3.860371e-02 4.466273e-02
#> [4,] -1.930243e-02 2.233203e-02
#> [5,] -9.651598e-03 1.116646e-02
#> [6,] -4.826038e-03 5.583506e-03
#> [7,] -2.413163e-03 2.791919e-03
#> [8,] -1.206665e-03 1.396057e-03
#> [9,] -6.033806e-04 6.980838e-04
#> [10,] -3.017172e-04 3.490731e-04
#> [11,] -1.508736e-04 1.745539e-04
#> [12,] -7.544503e-05 8.728646e-05
#> [13,] -3.772701e-05 4.364843e-05
#> [14,] -1.886594e-05 2.182703e-05
#> [15,] -9.434279e-06 1.091503e-05
#> [16,] -4.717841e-06 5.458327e-06
#>
#> $estimates
#> size_case size_rater p p_plus p_minus
#> [1,] 68 18 0.4984245 0.2794118 0.1029412
#>
#> $empirical
#> lower_bound mean upper_bound
#> [1,] 0.6029412 0.7944118 0.9705882
#> [2,] 0.5441176 0.6869118 0.8529412
#> [3,] 0.4558824 0.6277941 0.8088235
#> [4,] 0.4264706 0.5791176 0.7205882
#> [5,] 0.3970588 0.5411765 0.6764706
#> [6,] 0.3676471 0.5127941 0.6470588
#> [7,] 0.3529412 0.4879412 0.6323529
#> [8,] 0.3529412 0.4677941 0.5882353
#> [9,] 0.3529412 0.4416176 0.5588235
#> [10,] 0.3529412 0.4291176 0.5441176
#> [11,] 0.3529412 0.4136765 0.5294118
#> [12,] 0.3529412 0.4014706 0.4852941
#> [13,] 0.3529412 0.3889706 0.4852941
#> [14,] 0.3529412 0.3788235 0.4705882
#> [15,] 0.3529412 0.3677941 0.4558824
#> [16,] 0.3529412 0.3575000 0.3823529
#> [17,] 0.3529412 0.3529412 0.3529412
Some more details and examples can be found in vignettes of the package.
Reisenbichler, E. S., Han, G., Bellizzi, A., Bossuyt, V., Brock, J., Cole, K., Fadare, O., Hameed, O., Hanley, K., Harrison, B. T., Kuba, M. G., Ly, A., Miller, D., Podoll, M., Roden, A. C., Singh, K., Sanders, M. A., Wei, S., Wen, H., Pelekanou, V., Yaghoobi, V., Ahmed, F., Pusztai, L., and Rimm, D. L. (2020) “Prospective multi-institutional evaluation of pathologist assessment of PD-L1 assays for patient selection in triple negative breast cancer,” Mod Pathol, DOI: 10.1038/s41379-020-0544-x; PMID: 32300181.
Rimm, D. L., Han, G., Taube, J. M., Yi, E. S., Bridge, J. A., Flieder, D. B., Homer, R., West, W. W., Wu, H., Roden, A. C., Fujimoto, J., Yu, H., Anders, R., Kowalewski, A., Rivard, C., Rehman, J., Batenchuk, C., Burns, V., Hirsch, F. R., and Wistuba,, II (2017) “A Prospective, Multi-institutional, Pathologist-Based Assessment of 4 Immunohistochemistry Assays for PD-L1 Expression in Non-Small Cell Lung Cancer,” JAMA Oncol, 3(8), 1051-1058, DOI: 10.1001/jamaoncol.2017.0013, PMID: 28278348.