Design Parameter Optimization for Gold-Standard Non-Inferiority Trials.
OptimalGoldstandardDesigns 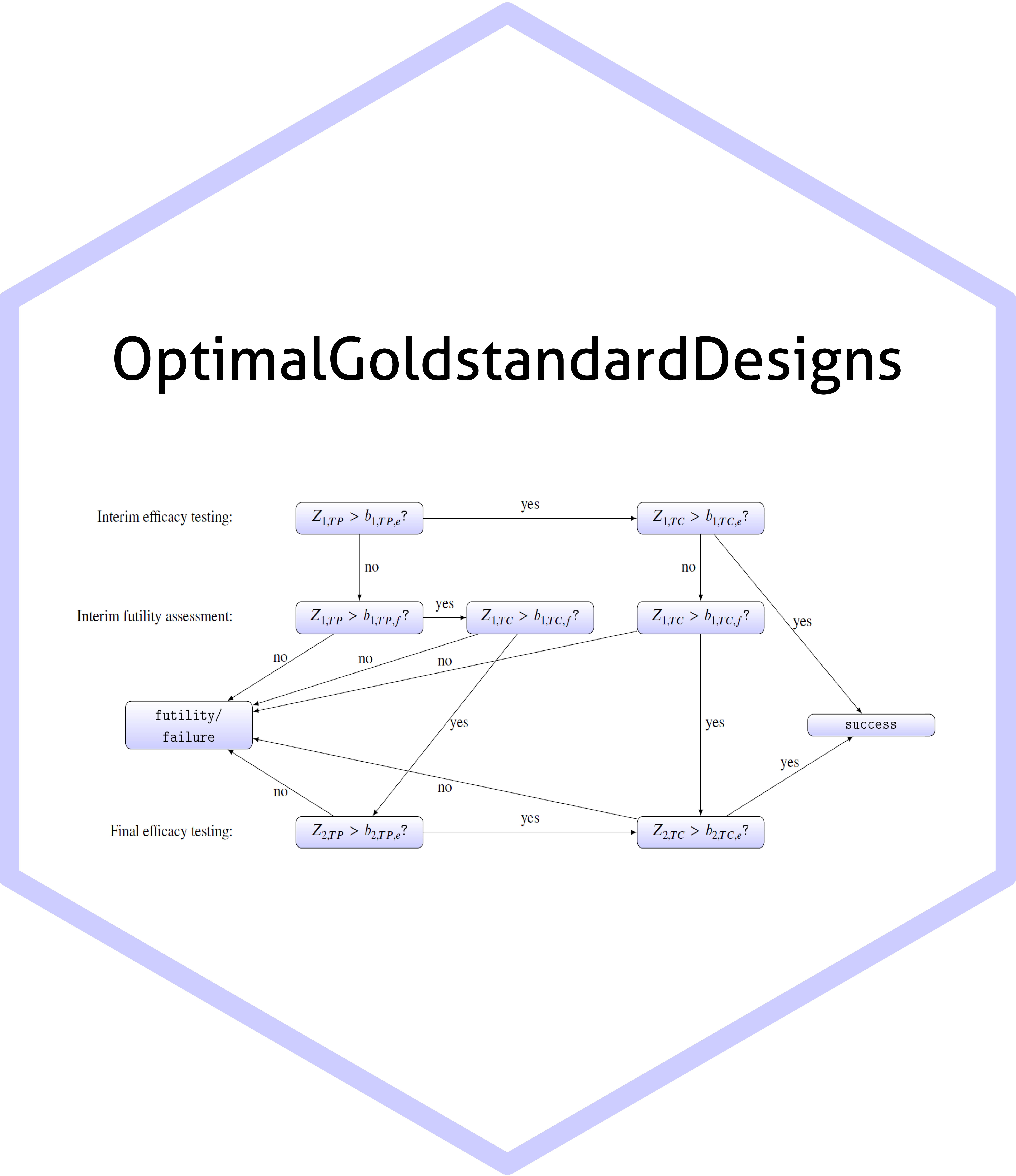
This package contains the code used in the calculations for our paper on the optimization of the two-stage group sequential three-arm gold-standard design for non-inferiority trials.
It allows for the simultaneous optimization of the allocation ratios for both stages (in two-stage designs), the efficacy boundaries, and the futility boundaries. The optimization is performed under type I and II error constraints and the objective function is customizable by the user. Methods to optimize two- and one-stage designs are available.
Installation
You can install the CRAN version of this package by typing the following command into your R console:
install.packages("OptimalGoldstandardDesigns")
You can install the GitHub Version by typing:
remotes::install_github("jan-imbi/OptimalGoldstandardDesigns")
You can also clone this repository directly from github. This will give you access to the /data/ subdirectory, which contains code to reproduce the examples from the paper.
Documentation
You can check out an online version of the documentation using this link.. In particular, you might be interested in reading the Usage guidance article.
Example
A one-stage desgin
library(OptimalGoldstandardDesigns)
optimize_design_onestage(
alpha = .025,
beta = .2,
alternative_TP = .4,
alternative_TC = 0,
Delta = .2,
print_progress = FALSE
)
#> Sample sizes (stage 1): T: 413, P: 125, C: 404
#> Efficacy boundaries (stage 1): Z_TP_e: 1.95996, Z_TC_e: 1.95996
#> Maximum overall sample size: 942
#> Placebo penalty at optimum (kappa * nP): 0.0
#> Objective function value: 942.0
#> Type I error for TP testing: 2.5%
#> Type I error for TC testing: 2.5%
#> Power: 80.2%
A two-stage design
optimize_design_twostage(
beta = 0.2,
alternative_TP = 0.4,
alternative_TC = 0,
Delta = 0.2,
print_progress = FALSE,
binding_futility = TRUE
)
#> Sample sizes (stage 1): T: 229, P: 90, C: 231
#> Sample sizes (stage 2): T: 217, P: 107, C: 199
#> Efficacy boundaries (stage 1): Z_TP_e: 2.04659, Z_TC_e: 2.29485
#> Futility boundaries (stage 1): Z_TP_f: 0.23336, Z_TC_f: 0.75795
#> Efficacy boundaries (stage 2): Z_TP_e: 2.40505, Z_TC_e: 2.04331
#> Inverse normal combination test weights (TP): w1: 0.68710, w2: 0.72656
#> Inverse normal combination test weights (TC): w1: 0.72466, w2: 0.68911
#> Maximum overall sample size: 1073
#> Expected sample size (H1): 768.5
#> Expected sample size (H0): 619.9
#> Expected placebo group sample size (H1): 100.2
#> Expected placebo group sample size (H0): 103.5
#> Objective function value: 768.5
#> (local) type I error for TP testing: 2.50%
#> (local) type I error for TC testing: 2.50%
#> Probability of futility stop (H1): 8.33%
#> Probability of futility stop (H0): 86.28%
#> Minimum conditional power: 34.17%
#> Power: 80.16%
#> Futility boundaries: binding
#> Futility testing method: always both futility tests
References
Meis, J, Pilz, M, Herrmann, C, Bokelmann, B, Rauch, G, Kieser, M. Optimization of the two-stage group sequential three-arm gold-standard design for non-inferiority trials. Statistics in Medicine. 2023; 42( 4): 536– 558. doi:10.1002/sim.9630.

