Analysis of Parent-Specific DNA Copy Numbers.
PSCBS: Analysis of Parent-Specific DNA Copy Numbers
The PSCBS package implements the parent-specific copy-number segmentation presented in Olshen et al. (2011). Package vignette 'Parent-specific copy-number segmentation using Paired PSCBS' provides a detailed introduction for running PSCBS segmentation. It's available as:
vignette("PairedPSCBS", package = "PSCBS")
Below is an excerpt of the example found in that vignette:
library(PSCBS)
## Get single-chromosome example data
data <- exampleData("paired.chr01")
str(data)
# ’data.frame’: 73346 obs. of 6 variables:
# $ chromosome: int 1 1 1 1 1 1 1 1 1 1 ...
# $ x : int 1145994 2224111 2319424 2543484 2926730 2941694 3084986 3155127..
# $ CT : num 1.625 1.071 1.406 1.18 0.856 ...
# $ betaT : num 0.757 0.771 0.834 0.778 0.229 ...
# $ CN : num 2.36 2.13 2.59 1.93 1.71 ...
# $ betaN : num 0.827 0.875 0.887 0.884 0.103 ...
## Drop total copy-number outliers
data <- dropSegmentationOutliers(data)
## Identify chromosome arms from data
gaps <- findLargeGaps(data, minLength = 1e+06)
knownSegments <- gapsToSegments(gaps)
## Parent-specific copy-number segmentation
fit <- segmentByPairedPSCBS(data, knownSegments = knownSegments)
## Get segments as a data.frame
segments <- getSegments(fit, simplify = TRUE)
segments
# chromosome tcnId dhId start end tcnNbrOfLoci tcnMean
# 1 1 1 1 554484 33414619 9413 1.381375
# 2 1 1 2 33414619 86993745 17433 1.378570
# 3 1 2 1 86993745 87005243 2 3.185100
# 4 1 3 1 87005243 119796080 10404 1.389763
# 5 1 3 2 119796080 119932126 72 1.470789
# 6 1 3 3 119932126 120992603 171 1.439620
# 7 1 4 1 120992604 141510002 0 NA
# 8 1 5 1 141510003 185527989 13434 2.065400
# 9 1 6 1 185527989 199122066 4018 2.707400
# 10 1 7 1 199122066 206512702 2755 2.586100
# 11 1 8 1 206512702 206521352 14 3.871900
# 12 1 9 1 206521352 247165315 15581 2.637500
# tcnNbrOfSNPs tcnNbrOfHets dhNbrOfLoci dhMean c1Mean c2Mean
# 1 2765 2765 2765 0.4868 0.3544608 1.0269140
# 2 4544 4544 4544 0.5185 0.3318907 1.0466792
# 3 0 0 0 NA NA NA
# 4 2777 2777 2777 0.5203 0.3333347 1.0564285
# 5 8 8 8 0.0767 0.6789900 0.7917995
# 6 52 52 52 0.5123 0.3510514 1.0885688
# 7 0 0 NA NA NA NA
# 8 3770 3770 3770 0.0943 0.9353164 1.1300836
# 9 1271 1271 1271 0.2563 1.0067467 1.7006533
# 10 784 784 784 0.2197 1.0089669 1.5771331
# 11 9 9 9 0.2769 1.3998854 2.4720146
# 12 4492 4492 4492 0.2290 1.0167563 1.6207438
## Plot copy-number tracks
plotTracks(fit)
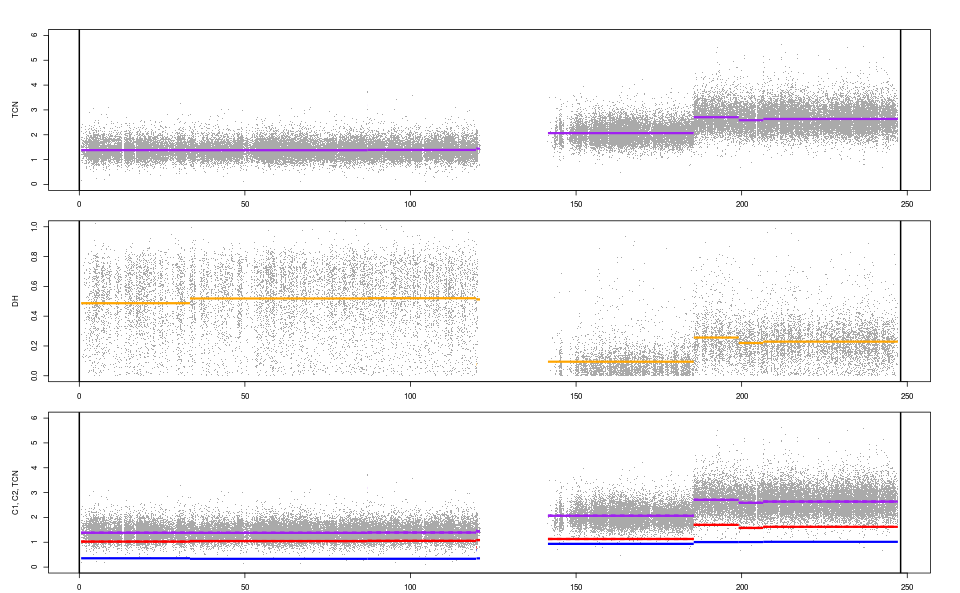
Parallel processing
The package supports segmentation of the chromosomes in parallel (asynchronously) via futures by adding the following
future::plan("multisession")
to the beginning of the PSCBS script. Everything else will work the same. To reset to non-parallel processing, use future::plan("sequential").
To configure this automatically whenever the package is loaded, see future vignette 'A Future for R: Controlling Default Future Strategy'.
References
Bengtsson H, Neuvial P, Speed TP. TumorBoost: Normalization of allele-specific tumor copy numbers from a single pair of tumor-normal genotyping microarrays, BMC Bioinformatics, 2010. DOI: 10.1186/1471-2105-11-245. PMID: 20462408, PMCID: PMC2894037
Olshen AB, Bengtsson H, Neuvial P, Spellman PT, Olshen RA, Seshan VA. Parent-specific copy number in paired tumor-normal studies using circular binary segmentation, Bioinformatics, 2011. DOI: 10.1093/bioinformatics/btr329. PMID: 21666266. PMCID: PMC3137217
Installation
R package PSCBS is available on CRAN and can be installed in R as:
# install.packages("BiocManager")
BiocManager::install(c("aroma.light", "DNAcopy"))
install.packages("PSCBS")
Pre-release version
To install the pre-release version that is available in Git branch develop on GitHub, use:
remotes::install_github("HenrikBengtsson/PSCBS", ref="develop")
This will install the package from source.