Axis Invariant Scatter Pie Plots.
PieGlyph 
PieGlyph is an R package aimed at replacing points in a plot with pie-chart glyphs, showing the relative proportions of different categories. The pie-chart glyphs are invariant to the axes and plot dimensions to prevent distortions when the plot dimensions are changed.
Installation
You can install the released version of PieGlyph from CRAN by running:
install.packages("PieGlyph")
Alternatively, you can install the development version of PieGlyph from GitHub with:
# install.packages("devtools")
devtools::install_github("rishvish/PieGlyph")
Examples
Load libraries
library(dplyr)
library(tidyr)
library(ggplot2)
library(PieGlyph)
library(ggiraph)
Simulate raw data
set.seed(123)
plot_data <- data.frame(response = rnorm(30, 100, 30),
system = 1:30,
group = sample(size = 30, x = c('G1', 'G2', 'G3'), replace = T),
A = round(runif(30, 3, 9), 2),
B = round(runif(30, 1, 5), 2),
C = round(runif(30, 3, 7), 2),
D = round(runif(30, 1, 9), 2))
The data has 30 observations and seven columns. response is a continuous variable measuring system output while system describes the 30 individual systems of interest. Each system is placed in one of three groups shown in group. Columns A, B, C, and D measure system attributes.
head(plot_data)
#> response system group A B C D
#> 1 83.18573 1 G1 5.80 1.57 4.78 8.31
#> 2 93.09468 2 G3 6.07 3.76 3.87 8.21
#> 3 146.76125 3 G1 6.60 3.48 5.01 3.19
#> 4 102.11525 4 G2 5.00 4.57 4.42 3.57
#> 5 103.87863 5 G1 5.93 3.69 5.60 8.89
#> 6 151.45195 6 G1 8.73 3.95 4.50 5.96
Create scatter plot with pie-charts
We can plot the outputs for each system as a scatterplot and replace the points with pie-chart glyphs showing the relative proportions of the four system attributes.
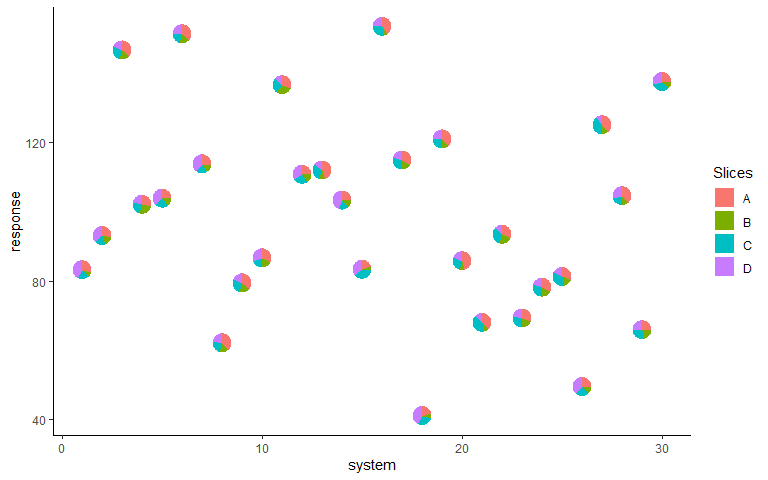
Basic plot
ggplot(data = plot_data, aes(x = system, y = response))+
geom_pie_glyph(slices = c('A', 'B', 'C', 'D'))+
theme_classic()
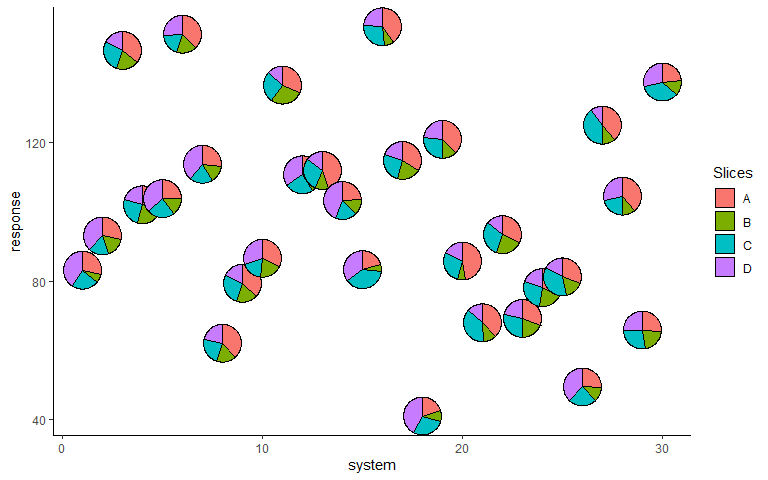
Change pie radius and border colour
ggplot(data = plot_data, aes(x = system, y = response))+
# Can also specify slices as column indices
geom_pie_glyph(slices = 4:7, colour = 'black', radius = 0.5)+
theme_classic()
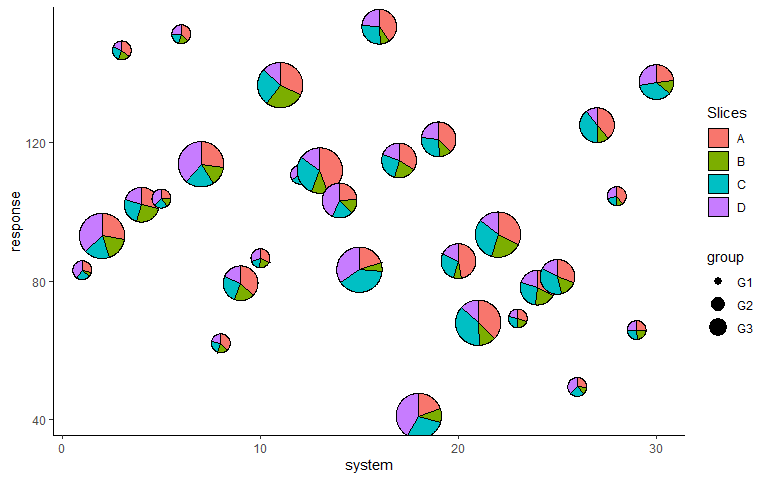
Map radius to a variable
p <- ggplot(data = plot_data, aes(x = system, y = response))+
geom_pie_glyph(aes(radius = group),
slices = c('A', 'B', 'C', 'D'),
colour = 'black')+
theme_classic()
p
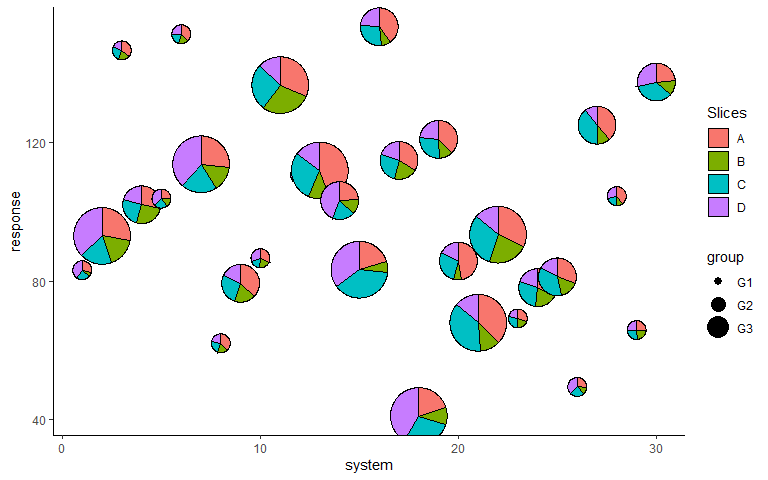
Adjust radius for groups
p <- p + scale_radius_manual(values = c(0.25, 0.5, 0.75), unit = 'cm')
p
Add custom labels
p <- p + labs(x = 'System', y = 'Response', fill = 'Attributes', radius = 'Group')
p
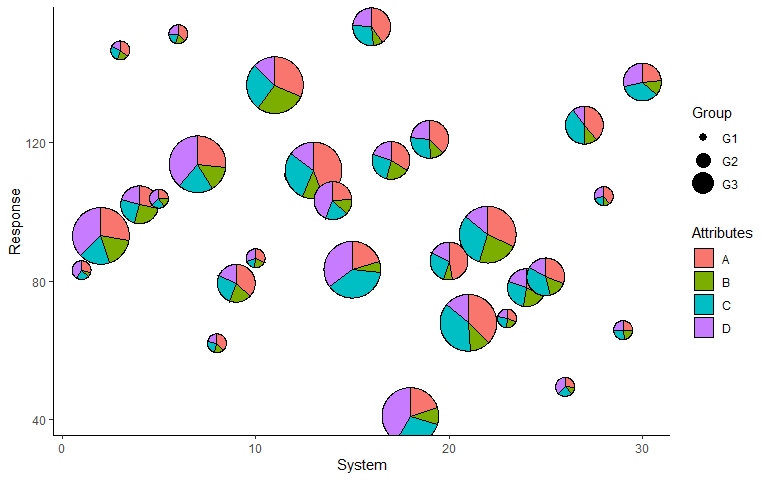
Change category colours
p + scale_fill_manual(values = c('#56B4E9', '#CC79A7', '#F0E442', '#D55E00'))
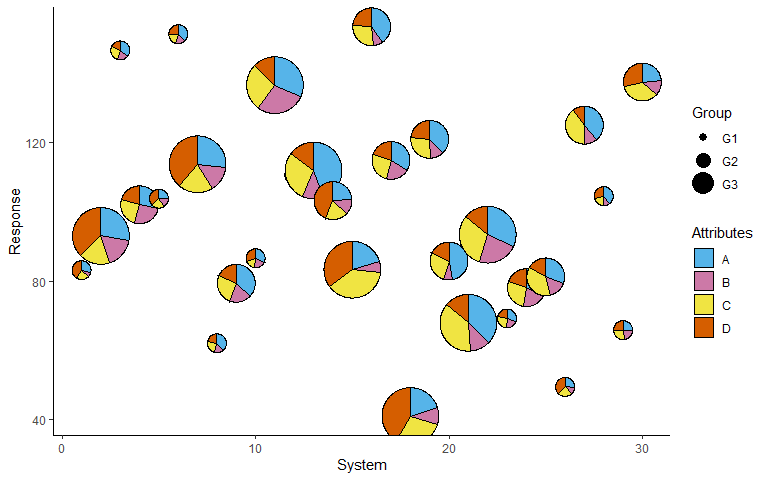
Alternative specification
The attributes can also be stacked into one column to generate the plot. This variant of the function is useful for situations when the data is in tidy format. See vignette('tidy-data') and vignette('pivot') for more information.
Stack the attributes in one column
plot_data_stacked <- plot_data %>%
pivot_longer(cols = c('A','B','C','D'),
names_to = 'Attributes',
values_to = 'values')
head(plot_data_stacked, 8)
#> # A tibble: 8 × 5
#> response system group Attributes values
#> <dbl> <int> <chr> <chr> <dbl>
#> 1 83.2 1 G1 A 5.8
#> 2 83.2 1 G1 B 1.57
#> 3 83.2 1 G1 C 4.78
#> 4 83.2 1 G1 D 8.31
#> 5 93.1 2 G3 A 6.07
#> 6 93.1 2 G3 B 3.76
#> 7 93.1 2 G3 C 3.87
#> 8 93.1 2 G3 D 8.21
Create plot
ggplot(data = plot_data_stacked, aes(x = system, y = response))+
# Along with categories column, values column is also needed now
geom_pie_glyph(slices = 'Attributes', values = 'values')+
theme_classic()
Interactive pie-chart glyphs
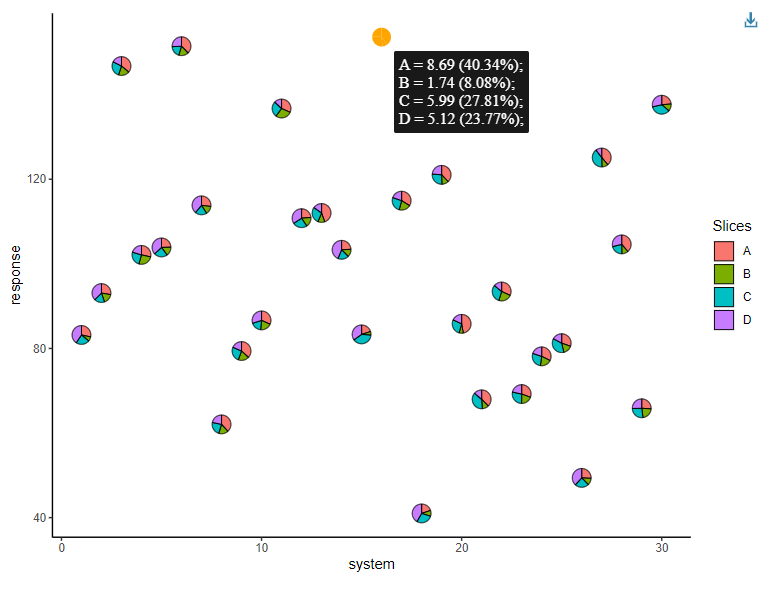
It is also possible to create interactive pie-chart scatterplots using the geom_pie_interactive function via the ggiraph framework.
Hovering over a pie-chart glyph will show a tooltip containing information about the raw counts and percentages of the categories (system attributes in this example) shown in the pie-charts. All additional features by ggiraph are also supported. See the ggiraph book and vignette("interactive-pie-glyphs") for more information.
plot_obj <- ggplot(data = plot_data)+
geom_pie_interactive(aes(x = system, y = response,
data_id = system),
slices = c("A", "B", "C", "D"),
colour = "black")+
theme_classic()
girafe(ggobj = plot_obj, height_svg = 6, width_svg = 8)