Heart Rate Turbulence Analysis.
RHRT
RHRT provides tools for a Heart Rate Turbulence analysis of RR interval data. It can either find the ventricular premature complexes (VPCs) via a set of filter rules or can use annotation data to only check the beats with the correct annotation. VPC snippets are filtered for validity and HRT parameters calculated.
In addition to standard calculation methods the package allows to modify the filter and calculation parameters. It is therefore not only helpful to identify HRT classes of measurements for risk assessment but also for assessment of the methodology itself.
For more information please check the vignette of the package. For more information about HRT have a look into the original publication by Schmidt et al. or our review with focus on the methodology.
Installation
You can install the released version of RHRT from CRAN with:
install.packages("RHRT")
And the development version from GitHub with:
install.packages("devtools") # if not already done
devtools::install_github("VBlesius/RHRT/RHRT")
Example
The general workflow of RHRT is to scan your interval data for HRT and check the results via HRT class and plot:
library(RHRT)
## scan your interval data and save the results as an HRTList
hrtl <- vectorToHRT(testdataLong)
## get the HRT class of your data
getResults(hrtl, type = "class")
#> [1] "HRT0"
## have a look at the data and the parameters
plot(hrtl)
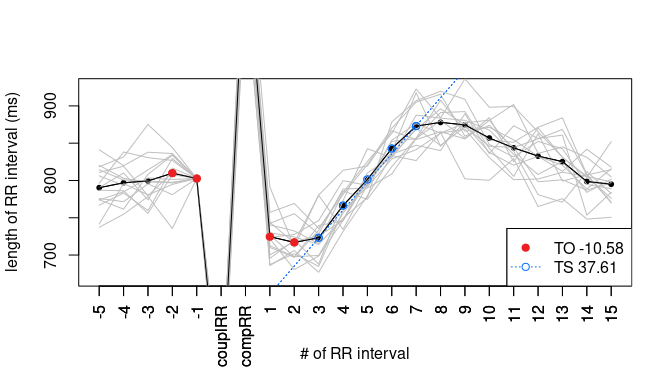
Data
Data to test the package can be found on Physionet. Via the WFDB Toolkit ECG data can be downloaded and/or converted, for example:
ann2rr -r chf2db/chf201 -a ecg -i s3 -w > ~/some/path/chf201.csv
Then load the data and use RHRT to find VPCSs:
chf201 <- read.table("~/some/path/chf201.csv")
hrtl <- RHRT::vectorToHRT(chf201[[1]]*1000, ann = chf201[[2]])
More example workflows can be found in the vignette.