Description
Implementations of Semi-Supervised Learning Approaches for Classification.
Description
A collection of implementations of semi-supervised classifiers and methods to evaluate their performance. The package includes implementations of, among others, Implicitly Constrained Learning, Moment Constrained Learning, the Transductive SVM, Manifold regularization, Maximum Contrastive Pessimistic Likelihood estimation, S4VM and WellSVM.
README.md
R Semi-Supervised Learning package
This R package provides implementations of several semi-supervised learning methods, in particular, our own work involving constraint based semi-supervised learning.
To cite the package, use either of these two references:
- Krijthe, J. H. (2016). RSSL: R package for Semi-supervised Learning. In B. Kerautret, M. Colom, & P. Monasse (Eds.), Reproducible Research in Pattern Recognition. RRPR 2016. Lecture Notes in Computer Science, vol 10214. (pp. 104–115). Springer International Publishing. https://doi.org/10.1007/978-3-319-56414-2_8. arxiv: https://arxiv.org/abs/1612.07993
- Krijthe, J.H. & Loog, M. (2015). Implicitly Constrained Semi-Supervised Least Squares Classification. In E. Fromont, T. de Bie, & M. van Leeuwen, eds. 14th International Symposium on Advances in Intelligent Data Analysis XIV (Lecture Notes in Computer Science Volume 9385). Saint Etienne. France, pp. 158-169.
Installation Instructions
This package available on CRAN. The easiest way to install the package is to use:
install.packages("RSSL")
To install the latest version of the package using the devtools package:
library(devtools)
install_github("jkrijthe/RSSL")
Usage
After installation, load the package as usual:
library(RSSL)
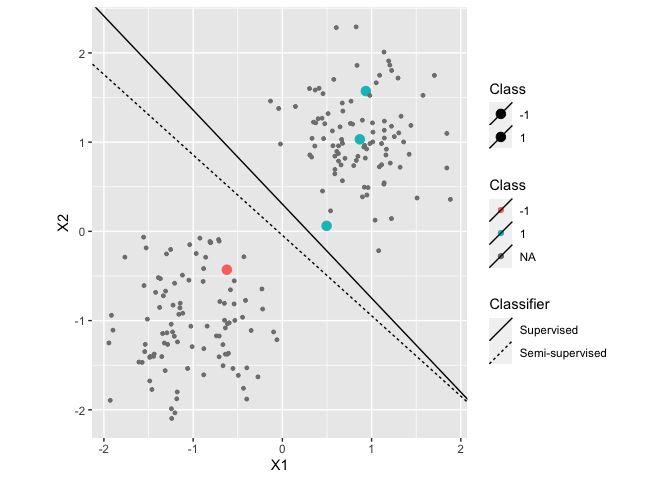
The following code generates a simple dataset, trains a supervised and two semi-supervised classifiers and evaluates their performance:
library(dplyr,warn.conflicts = FALSE)
library(ggplot2,warn.conflicts = FALSE)
set.seed(2)
df <- generate2ClassGaussian(200, d=2, var = 0.2, expected=TRUE)
# Randomly remove labels
df <- df %>% add_missinglabels_mar(Class~.,prob=0.98)
# Train classifier
g_nm <- NearestMeanClassifier(Class~.,df,prior=matrix(0.5,2))
g_self <- SelfLearning(Class~.,df,
method=NearestMeanClassifier,
prior=matrix(0.5,2))
# Plot dataset
df %>%
ggplot(aes(x=X1,y=X2,color=Class,size=Class)) +
geom_point() +
coord_equal() +
scale_size_manual(values=c("-1"=3,"1"=3), na.value=1) +
geom_linearclassifier("Supervised"=g_nm,
"Semi-supervised"=g_self)
# Evaluate performance: Squared Loss & Error Rate
mean(loss(g_nm,df))
mean(loss(g_self,df))
mean(predict(g_nm,df)!=df$Class)
mean(predict(g_self,df)!=df$Class)
Acknowledgement
Work on this package was supported by Project 23 of the Dutch national program COMMIT.