Concentration-Response Data Analysis using Curvep.
Overview
The package provides an R interface for processing concentration-response datasets using Curvep, a response noise filtering algorithm. The algorithm was described in the publications (Sedykh A et al. (2011) doi:10.1289/ehp.1002476 and Sedykh A (2016) doi:10.1007/978-1-4939-6346-1_14).
Other parametric fitting approaches (e.g., Hill equation) are also adopted for ease of comparison. 3-parameter Hill equation from original tcpl package (Filer DL et al., doi:10.1093/bioinformatics/btw680) and 4-parameter Hill equation from Curve Class2 approach (Wang Y et al., doi:10.2174/1875397301004010057) are available.
Also, methods for calculating the confidence interval around the activity metrics are also provided. The methods are based on the bootstrap approach to simulate the datasets (Hsieh J-H et al. doi:10.1093/toxsci/kfy258). The simulated datasets can be used to derive the baseline noise threshold in an assay endpoint. This threshold is critical in the toxicological studies to derive the point-of-departure (POD).
Installation
# the development version from GitHub:
# install.packages("devtools")
devtools::install_github("moggces/Rcurvep")
devtools::install_github("moggces/Rcurvep", dependencies = TRUE, build_vignettes = TRUE)
Package structure
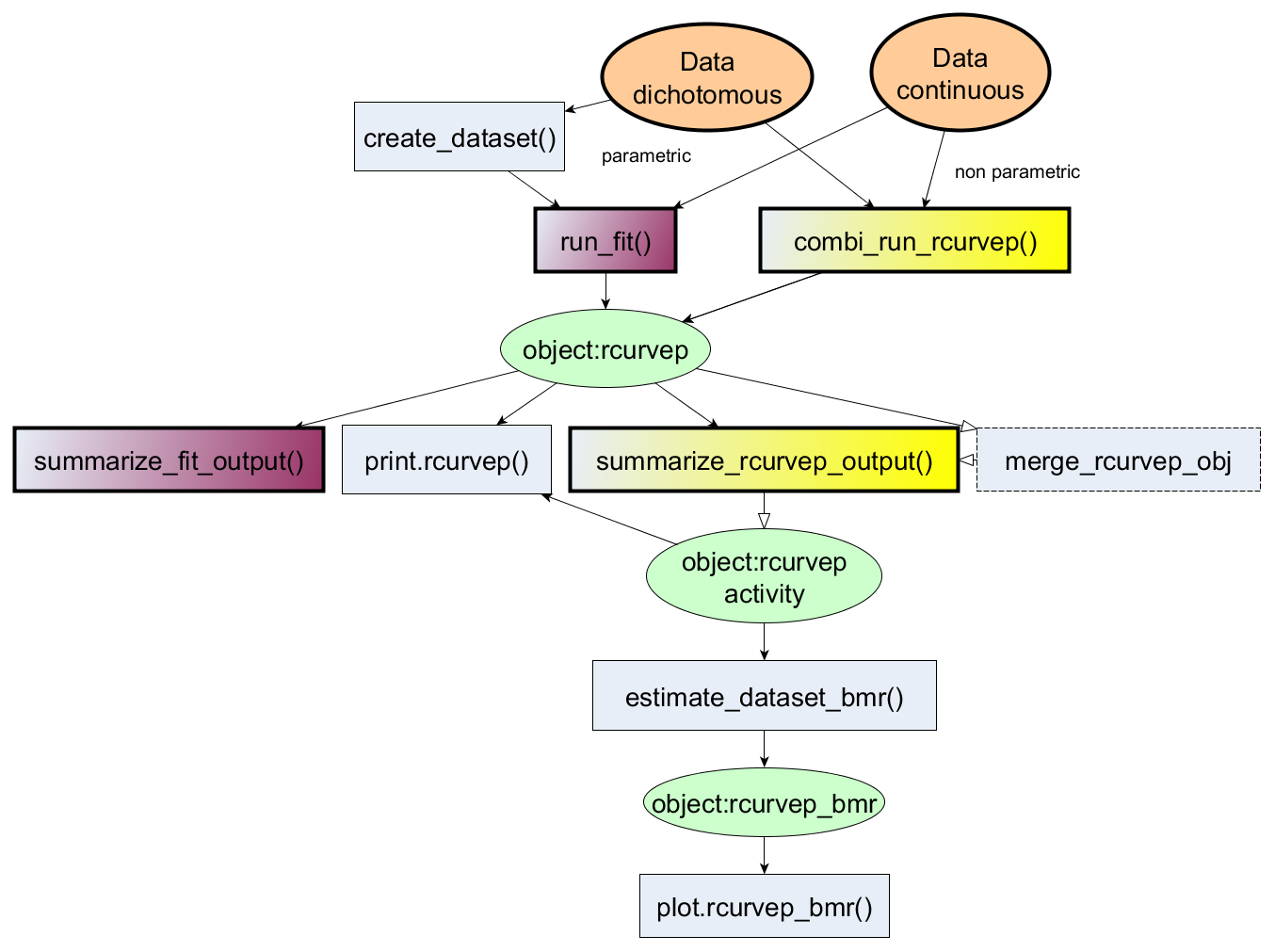
Usage
Run analysis
library(Rcurvep)
data("zfishbeh")
out_curvep <- combi_run_rcurvep(zfishbeh, TRSH = 30) # using Curvep with BMR = 30
out_fit1 <- run_fit(zfishbeh, modls = "cc2") # using Curve Class2 4-parameter hill
out_fit2 <- run_fit(zfishbeh, modls = c("cnst", "hill")) # using tcpl 3-parameter hill + constant model
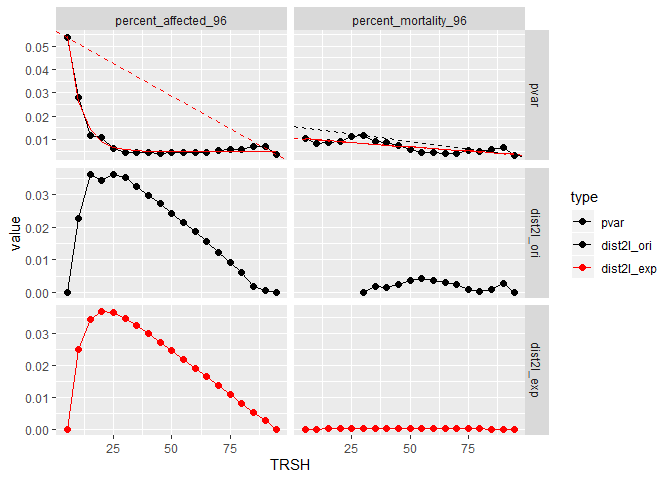
Find BMR
data("zfishdev_act")
out_bmr <- estimate_dataset_bmr(zfishdev_act)
## $`1`
More Usage
To learn more about Rcurvep, start with the vignettes: browseVignettes(package = "Rcurvep")