Next Eigenvalue Sufficiency Test.
Rnest: An R package for the Next Eigenvalue Sufficiency Test
The library Rnest offers the Next Eigenvalue Sufficiency Tests (NEST) (Achim, 2017, 2020) to determine the number of dimensions in exploratory factor analysis. It provides a main function nest() to carry the analysis and a plot() function.
There is also many examples of correlation matrices available with the packages and other stopping rules as well, such as pa() for parellel analysis.
Installation
The development version can be accessed through GitHub:
remotes::install_github(repo = "quantmeth/Rnest")
library(Rnest)
Examples
Here is an example using the ex_4factors_corr correlation matrix from the Rnest library. The factor structure is
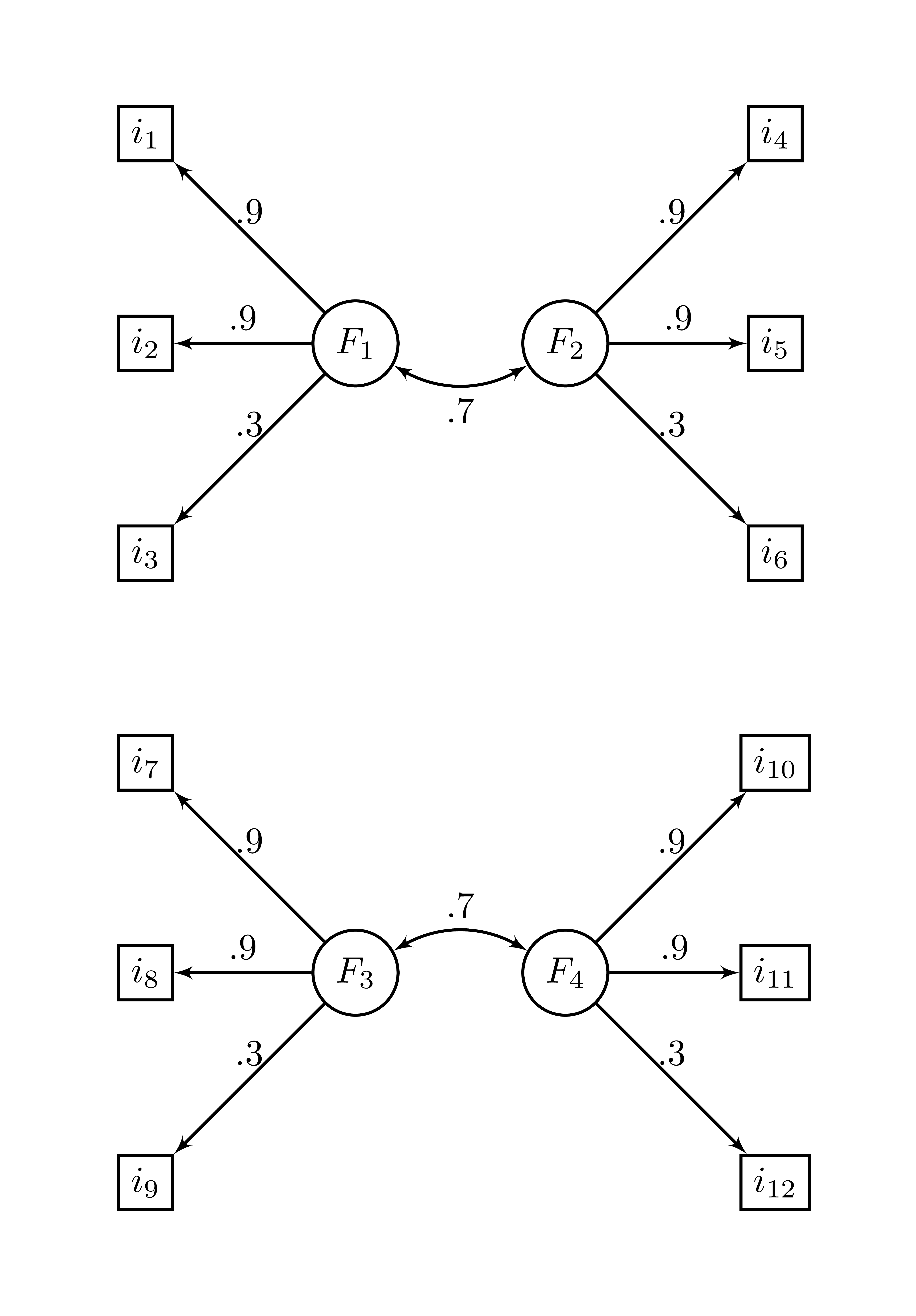
and the correlation matrix is
$$\begin{bmatrix} 1&0.81&0.27&0.567&0.567&0.189&0&0&0&0&0&0 \
0.81&1&0.27&0.567&0.567&0.189&0&0&0&0&0&0 \
0.27&0.27&1&0.189&0.189&0.063&0&0&0&0&0&0 \
0.567&0.567&0.189&1&0.81&0.27&0&0&0&0&0&0 \
0.567&0.567&0.189&0.81&1&0.27&0&0&0&0&0&0 \
0.189&0.189&0.063&0.27&0.27&1&0&0&0&0&0&0 \
0&0&0&0&0&0&1&0.81&0.27&0.567&0.567&0.189 \
0&0&0&0&0&0&0.81&1&0.27&0.567&0.567&0.189 \
0&0&0&0&0&0&0.27&0.27&1&0.189&0.189&0.063 \
0&0&0&0&0&0&0.567&0.567&0.189&1&0.81&0.27 \
0&0&0&0&0&0&0.567&0.567&0.189&0.81&1&0.27 \
0&0&0&0&0&0&0.189&0.189&0.063&0.27&0.27&1 \
\end{bmatrix}$$
From ex_4factors_corr, we can easily generate random data using the MASS packages (Venables & Ripley, 2002).
set.seed(1)
mydata <- MASS::mvrnorm(n = 2500,
mu = rep(0, ncol(ex_4factors_corr)),
Sigma = ex_4factors_corr)
We can then carry NEST.
res <- nest(mydata)
res
## At 95% confidence, Nest Eigenvalue Sufficiency Test (NEST) suggests 4 factors.
The first output tells hom many factors NEST suggest. We can also consult the summary with
summary(res)
##
## nest 0.0.0.1 ended normally
##
## Estimator ML
## Number of model parameters 66
## Resampling 1000
## Sample size 2500
## Stopped at 5
##
##
## Probabilities of factors
## Factor Eigenvalue Prob
## F1 3.228 < .001
## F2 3.167 < .001
## F3 1.007 .009
## F4 0.972 .009
## F5 0.860 .735
##
##
## At 95% confidence, Nest Eigenvalue Sufficiency Test (NEST) suggests 4 factors.
## Try plot(nest()) to see a graphical representation of the results.
##
We can visualize the results using the generic function plot() using the nest() output.
plot(res)
Scree plot of NEST
The above figure shows the empirical eigenvalues in blue and the 95th percentile of the sampled eigenvalues.
It is also possible to use a correlation matrix directly. A sample size, n must be supplied.
nest(ex_4factors_corr, n = 240)
## At 95% confidence, Nest Eigenvalue Sufficiency Test (NEST) suggests 2 factors.
The nest() function can use with many $\alpha$ values if desired.
res <- nest(ex_4factors_corr, n = 120, alpha = c(.01,.025,.05,.1))
plot(res)
Scree plot of NEST with many $\alpha$
How to cite
Caron, P.-O. (2023). Rnest: An R package for the Next Eigenvalue Sufficiency Test. https://github.com/quantmeth/Rnest
References
Achim, A. (2017). Testing the number of required dimensions in exploratory factor analysis. The Quantitative Methods for Psychology, 13(1), 64–74. https://doi.org/10.20982/tqmp.13.1.p064
Achim, A. (2020). Esprit et enjeux de l’analyse factorielle exploratoire. The Quantitative Methods for Psychology, 16(4), 213–247. https://doi.org/10.20982/tqmp.16.4.p213
Venables, W. N., & Ripley, B. D. (2002). Modern applied statistics with S. Springer. https://www.stats.ox.ac.uk/pub/MASS4/