Fitting Shared Atoms Nested Models via Markov Chains Monte Carlo.
SANple v0.1.1
The goal of SANple is to estimate Bayesian nested mixture models via MCMC methods. Specifically, the package implements the common atoms model (Denti et al., 2023), its finite version (D’Angelo et al., 2023), and a hybrid finite-infinite model (D’Angelo and Denti, 2024+). All models use Gaussian mixtures with a normal-inverse-gamma prior distribution on the parameters. Additional functions are provided to help analyzing the results of the fitting procedure.
Installation
You can install the development version of SANple from GitHub with:
# install.packages("devtools")
devtools::install_github("laura-dangelo/SANple")
Example
This is a basic example which shows you how to solve a common problem:
library(SANple)
#> Loading required package: scales
#> Loading required package: RColorBrewer
## basic example code
set.seed(123)
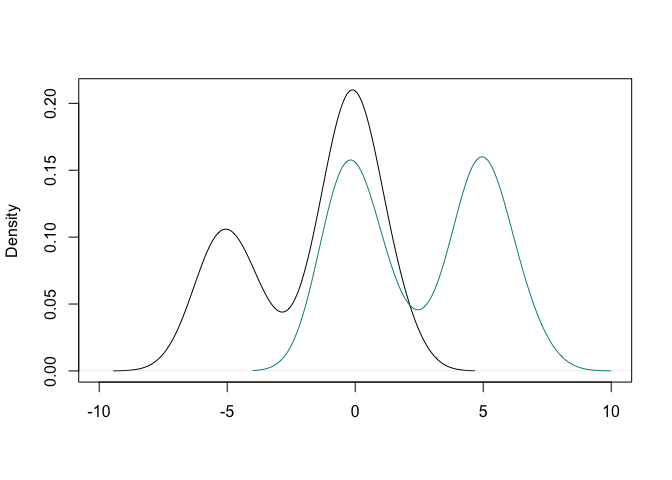
y <- c(rnorm(50,-5,1), rnorm(170,0,1),rnorm(70,5,1))
g <- c(rep(1,150), rep(2, 140))
plot(density(y[g==1]), xlim = c(-10,10), main = "", xlab = "")
lines(density(y[g==2]), col = "cyan4")
out <- sample_fiSAN(nrep = 3000, burn = 1000, y = y, group = g, beta = 1)
out
#>
#> MCMC result of fiSAN model
#> -----------------------------------------------
#> Model estimated on 290 total observations and 2 groups
#> Total MCMC iterations: 3000
#> maxL: 50 - maxK: 50
#> Elapsed time: 1.625 secs
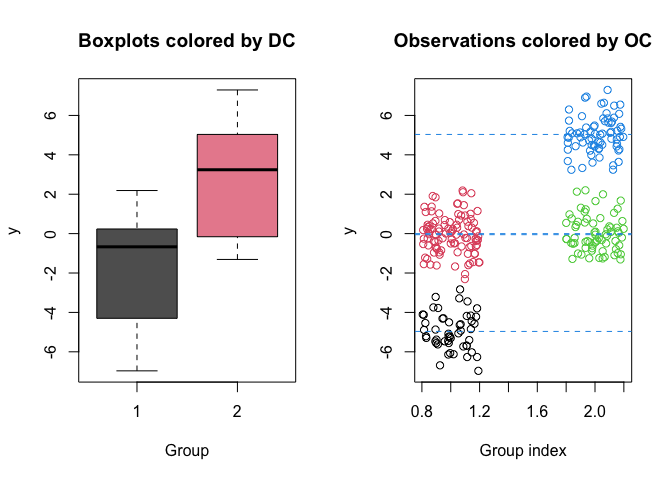
clusters <- estimate_clusters(out)
clusters
#>
#> Summary of the estimated observational and distributional clusters
#>
#> ----------------------------------
#> Estimated number of observational clusters: 4
#> Estimated number of distributional clusters: 2
#> ----------------------------------
#>
#> Distributional cluster 1
#> post_mean post_var
#> 1 -4.96559645 0.8572352
#> 2 -0.05374608 0.9306906
#>
#> Distributional cluster 2
#> post_mean post_var
#> 3 -0.00160923 0.7737203
#> 4 5.03452815 0.8314760
plot(out, estimated_clusters = clusters)
References
D’Angelo, L., Canale, A., Yu, Z., Guindani, M. (2023). Bayesian nonparametric analysis for the detection of spikes in noisy calcium imaging data. Biometrics 79(2), 1370–1382.
D’Angelo, L., and Denti, F. (2024+). A finite-infinite shared atoms nested model for the Bayesian analysis of large grouped data sets. Working paper, 1–34
Denti, F., Camerlenghi, F., Guindani, M., Mira, A., 2023. A Common Atoms Model for the Bayesian Nonparametric Analysis of Nested Data. Journal of the American Statistical Association. 118(541), 405–416.
