Simulation of Correlated Data with Multiple Variable Types.
SimMultiCorrData
The goal of SimMultiCorrData is to generate continuous (normal or non-normal), binary, ordinal, and count (Poisson or Negative Binomial) variables with a specified correlation matrix. It can also produce a single continuous variable. This package can be used to simulate data sets that mimic real-world situations (i.e. clinical data sets, plasmodes, as in Vaughan et al., 2009). All variables are generated from standard normal variables with an imposed intermediate correlation matrix. Continuous variables are simulated by specifying mean, variance, skewness, standardized kurtosis, and fifth and sixth standardized cumulants using either Fleishman's Third-Order or Headrick's Fifth-Order Polynomial Transformation. Binary and ordinal variables are simulated using a modification of GenOrd::ordsample. Count variables are simulated using the inverse cdf method. There are two simulation pathways which differ primarily according to the calculation of the intermediate correlation matrix Sigma. In Correlation Method 1, the intercorrelations involving count variables are determined using a simulation based, logarithmic correlation correction (adapting Yahav and Shmueli's 2012 method). In Correlation Method 2, the count variables are treated as ordinal (adapting Barbiero and Ferrari's 2015 modification of GenOrd). There is an optional error loop that corrects the final correlation matrix to be within a user-specified precision value. The package also includes functions to calculate standardized cumulants for theoretical distributions or from real data sets, check if a target correlation matrix is within the possible correlation bounds (given the distributions of the simulated variables), summarize results (numerically or graphically), to verify valid power method pdfs, and to calculate lower standardized kurtosis bounds.
There are several vignettes which accompany this package that may help the user understand the simulation and analysis methods.
Benefits of SimMultiCorrData and Comparison to Other Packages describes some of the ways
SimMultiCorrDataimproves upon other simulation packages.Variable Types describes the different types of variables that can be simulated in
SimMultiCorrData.Function by Topic describes each function, separated by topic.
Comparison of Correlation Method 1 and Correlation Method 2 describes the two simulation pathways that can be followed.
Overview of Error Loop details the algorithm involved in the optional error loop that improves the accuracy of the simulated variables' correlation matrix.
Overall Workflow for Data Simulation gives a step-by-step guideline to follow with an example containing continuous (normal and non-normal), binary, ordinal, Poisson, and Negative Binomial variables. It also demonstrates the use of the standardized cumulant calculation function, correlation check functions, the lower kurtosis boundary function, and the plotting functions.
Comparison of Simulation Distribution to Theoretical Distribution or Empirical Data gives a step-by-step guideline for comparing a simulated univariate continuous distribution to the target distribution with an example.
Using the Sixth Cumulant Correction to Find Valid Power Method Pdfs demonstrates how to use the sixth cumulant correction to generate a valid power method pdf and the effects this has on the resulting distribution.
Installation instructions
SimMultiCorrData can be installed using the following code:
## from GitHub
install.packages("devtools")
devtools::install_github("AFialkowski/SimMultiCorrData")
## from CRAN
install.packages("SimMultiCorrData")
Example
This is a basic example which shows you how to solve a common problem: Compare a simulated exponential(mean = 2) variable to the theoretical exponential(mean = 2) density.
Step 1: Obtain the standardized cumulants
In R, the exponential parameter is rate <- 1/mean.
library(SimMultiCorrData)
stcums <- calc_theory(Dist = "Exponential", params = 0.5)
stcums
#> mean sd skew kurtosis fifth sixth
#> 2 2 2 6 24 120
Step 2: Simulate the variable
Note that calc_theory returns the standard deviation, not the variance. The simulation functions require variance as the input.
H_exp <- nonnormvar1("Polynomial", means = stcums[1], vars = stcums[2]^2,
skews = stcums[3], skurts = stcums[4],
fifths = stcums[5], sixths = stcums[6], Six = NULL,
cstart = NULL, n = 10000, seed = 1234)
#> Constants: Distribution 1
#>
#> Constants calculation time: 0 minutes
#> Total Simulation time: 0.001 minutes
names(H_exp)
#> [1] "constants" "continuous_variable" "summary_continuous"
#> [4] "summary_targetcont" "sixth_correction" "valid.pdf"
#> [7] "Constants_Time" "Simulation_Time"
# Look at constants
H_exp$constants
#> c0 c1 c2 c3 c4 c5
#> 1 -0.3077396 0.8005605 0.318764 0.03350012 -0.00367481 0.0001587077
# Look at summary
round(H_exp$summary_continuous[, c("Distribution", "mean", "sd", "skew",
"skurtosis", "fifth", "sixth")], 5)
#> Distribution mean sd skew skurtosis fifth sixth
#> X1 1 1.99987 2.0024 2.03382 6.18067 23.74145 100.3358
Step 3: Determine if the constants generate a valid power method pdf
H_exp$valid.pdf
#> [1] "TRUE"
Step 4: Select a critical value
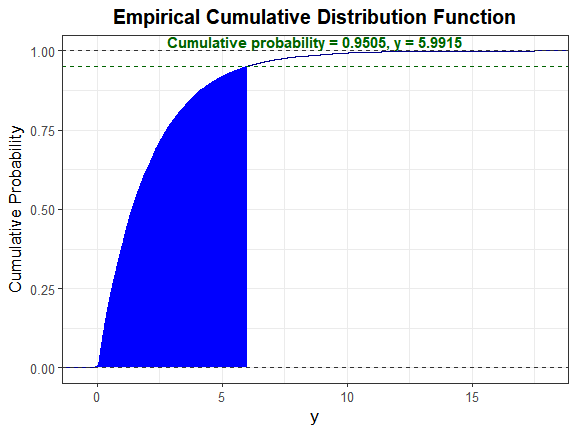
Let alpha = 0.05.
y_star <- qexp(1 - 0.05, rate = 0.5) # note that rate = 1/mean
y_star
#> [1] 5.991465
Step 5: Solve for $\Large z'$
Since the exponential(2) distribution has a mean and standard deviation equal to 2, solve $\Large 2 * p(z') + 2 - y_star = 0$ for $\Large z'$. Here, $\Large p(z') = c0 + c1 * z' + c2 * z'^2 + c3 * z'^3 + c4 * z'^4 + c5 * z'^5$.
f_exp <- function(z, c, y) {
return(2 * (c[1] + c[2] * z + c[3] * z^2 + c[4] * z^3 + c[5] * z^4 +
c[6] * z^5) + 2 - y)
}
z_prime <- uniroot(f_exp, interval = c(-1e06, 1e06),
c = as.numeric(H_exp$constants), y = y_star)$root
z_prime
#> [1] 1.644926
Step 6: Calculate $\Large \Phi(z')$
1 - pnorm(z_prime)
#> [1] 0.04999249
This is approximately equal to the alpha value of 0.05, indicating the method provides a good approximation to the actual distribution.
Step 7: Plot graphs
plot_sim_pdf_theory(sim_y = as.numeric(H_exp$continuous_variable[, 1]),
overlay = TRUE, Dist = "Exponential", params = 0.5)
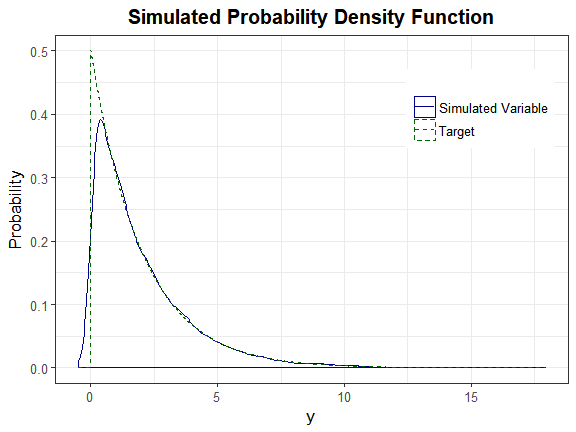
We can also plot the empirical cdf and show the cumulative probability up to y_star.
plot_sim_cdf(sim_y = as.numeric(H_exp$continuous_variable[, 1]),
calc_cprob = TRUE, delta = y_star)
Calculate descriptive statistics.
stats_pdf(c = H_exp$constants[1, ], method = "Polynomial", alpha = 0.025,
mu = 2, sigma = 2)
#> trimmed_mean median mode max_height
#> 1.858381 1.384521 0.104872 1.094213