Fit Spatial Generalized Extreme Value Models.
SpatialGEV
Meixi Chen, Martin Lysy, Reza Ramezan
Description
A fast Bayesian inference method for spatial random effects modelling of weather extremes. The latent spatial variables are efficiently marginalized by a Laplace approximation using the TMB library, which leverages efficient automatic differentiation in C++. The models are compiled in C++, whereas the optimization step is carried out in R. With this package, users can fit spatial GEV models with different complexities to their dataset without having to formulate the model using C++. This package also offers a method to sample from the approximate posterior distributions of both fixed and random effects, which will be useful for downstream analysis.
Installation
Before installing SpatialGEV, make sure you have TMB installed following the instructions here.
SpatialGEV uses several functions from the INLA package for SPDE approximation to the Matérn covariance as well as mesh creation on the spatial domain. If the user would like to use the SPDE method (i.e. kernel="spde" in spatialGEV_fit()), please first install package INLA. Since INLA is not on CRAN, it needs to be downloaded following their instruction here.
To download the stable version of this package, run
install.packages("SpatialGEV")
To download the development version of this package, run
devtools::install_github("meixichen/SpatialGEV")
Example
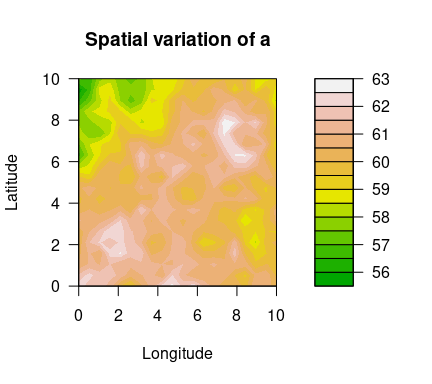
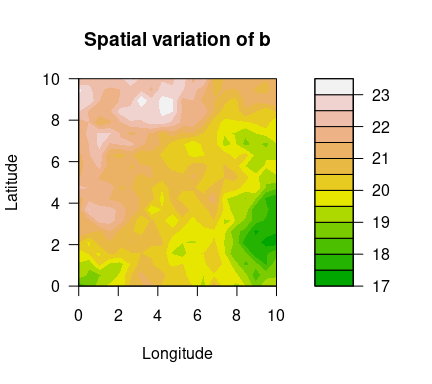
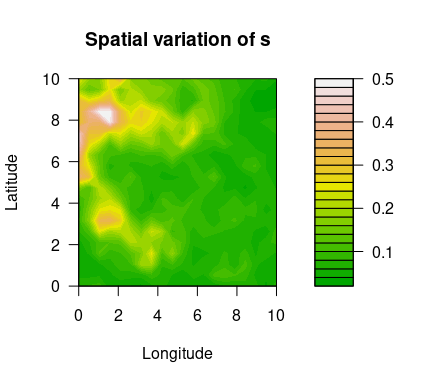
Using the simulated data set simulatedData2 provided in the package, we demonstrate how to use this package. Spatial variation of the GEV parameters are plotted below.
library(SpatialGEV)
# GEV parameters simulated from Gaussian random fields
a <- simulatedData2$a # location
logb <- simulatedData2$logb # log scale
logs <- simulatedData2$logs # log shape
locs <- simulatedData2$locs # coordinate matrix
n_loc <- nrow(locs) # number of locations
y <- Map(evd::rgev, n=sample(50:70, n_loc, replace=TRUE),
loc=a, scale=exp(logb), shape=exp(logs)) # observations
filled.contour(unique(locs$x), unique(locs$y), matrix(a, ncol=sqrt(n_loc)),
color.palette = terrain.colors, xlab="Longitude", ylab="Latitude",
main="Spatial variation of a",
cex.lab=1,cex.axis=1)
filled.contour(unique(locs$x), unique(locs$y), matrix(exp(logb), ncol=sqrt(n_loc)),
color.palette = terrain.colors, xlab="Longitude", ylab="Latitude",
main="Spatial variation of b",
cex.lab=1,cex.axis=1)
filled.contour(unique(locs$x), unique(locs$y), matrix(exp(logs), ncol=sqrt(n_loc)),
color.palette = terrain.colors, xlab="Longitude", ylab="Latitude",
main="Spatial variation of s",
cex.lab=1,cex.axis=1)
To fit a GEV-GP model to the simulated data, use the spatialGEV_fit() function. We use random="abs" to indicate that all three GEV parameters are treated as random effects. The shape parameter s is constrained to be positive (log transformed) by specifying reparam_s="positive". The covariance kernel function used here is the SPDE-approximated Matérn kernel kernel="spde". Initial parameter values are passed to init_param using a list.
fit <- spatialGEV_fit(data = y, locs = locs, random = "abs",
init_param = list(a = rep(60, n_loc),
log_b = rep(2,n_loc),
s = rep(-3,n_loc),
beta_a = 60, beta_b = 2, beta_s = -2,
log_sigma_a = 1.5, log_kappa_a = -2,
log_sigma_b = 1.5, log_kappa_b = -2,
log_sigma_s = -1, log_kappa_s = -2),
reparam_s = "positive", kernel="spde", silent = TRUE)
class(fit)
#> [1] "spatialGEVfit"
print(fit)
#> Model fitting took 27.9517922401428 seconds
#> The model has reached relative convergence
#> The model uses a spde kernel
#> Number of fixed effects in the model is 9
#> Number of random effects in the model is 1308
#> Hessian matrix is positive definite. Use spatialGEV_sample to obtain posterior samples
Posterior samples of the random and fixed effects are drawn using spatialGEV_sample(). Specify observation=TRUE if we would also like to draw from the posterior predictive distribution.
sam <- spatialGEV_sample(model = fit, n_draw = 1000, observation = T)
print(sam)
#> The samples contains 1000 draws of 1209 parameters
#> The samples contains 1000 draws of response at 400 locations
#> Use summary() to obtain summary statistics of the samples
To get summary statistics of the posterior samples, use summary() on the sample object.
pos_summary <- summary(sam)
pos_summary$param_summary[1:5,]
#> 2.5% 25% 50% 75% 97.5% mean
#> a1 59.98220 61.58412 62.39717 63.23451 64.98185 62.40852
#> a2 60.39339 61.75672 62.55552 63.30514 64.95137 62.55192
#> a3 60.01955 61.28410 61.98922 62.71080 64.13772 61.98618
#> a4 59.57800 60.92354 61.66539 62.39592 63.81774 61.66007
#> a5 59.55668 60.86209 61.54278 62.21564 63.68217 61.55340
pos_summary$y_summary[1:5,]
#> 2.5% 25% 50% 75% 97.5% mean
#> y1 39.43693 56.12927 70.41943 87.22232 140.3342 74.81315
#> y2 37.09989 55.68274 67.98861 87.42057 148.5369 74.42377
#> y3 38.46096 55.07948 69.19833 87.29214 152.6150 74.99553
#> y4 38.34585 56.38710 68.56856 86.17946 143.4025 73.64138
#> y5 37.81648 55.14434 69.23200 85.81161 143.7851 74.12148
TODO
[ ] Consider a shorter name, e.g.,
sgev_*, thanspatialGEV_*.[ ] Argument
init_paramsadds a lot of complexity tospatialGEV_fit(). Perhaps this function can be broken down into two parts:spatialGEV_adfun()which returns theadfunobject, and thenspatialGEV_fit()which does the fitting. The advantage is that theadfun$env$parametersobject tells you the dimension of the parameter values, which can be useful for initialization. Also, note thatadfun$env$dataobject contains the entire data list for browsing.[ ] Write some tests for the
spdekernel. Construct the sparse precision matrix usingget_spde_prec(), invert it usingMatrix::solve(), apply the scale factor, and pass as variance todmvnorm().