Spatial Structural Diversity Quantification in Raster Data.
StrucDiv
The R package StrucDiv contains methods to quantify scale-specific and multi-scale spatial structural diversity. Spatial structural diversity refers to the spatial, i.e. horizontal, configuration of landscape elements. Hereafter, ‘spatial structural diversity’ will be used synonymous to ‘structural diversity.’ The StrucDiv package provides methods to quantify structural diversity in continuous raster data. With these methods, structural diversity features can be detected. Structural diversity features have also been called latent landscape features. Typical structural diversity features include patches and linear landscape features and reveal transition zones between spatial regimes. Structural diversity can also be quantified in categorical raster data, required method specifications are described in Schuh, Santos, de Jong, et al. (2022). Structure is quantified by considering the spatial arrangement of pixel values as pairs, based on Haralick, Shanmugam, and Dinstein (1973). Structural diversity can be quantified on different spatial scales, whereby pixel resolution is an input to these methods, and different extents can be implemented. Scale-specific structural diversity can be quantified, either of the domain, or of smaller areas therein. The latter is done with a moving window approach. Multi-scale structural diversity is quantified by combining information from different scales (i.e. extents), as further described below. The methods are described in detail in Schuh, Santos, de Jong, et al. (2022) and in Schuh, Santos, Schaepman, et al. (2022), and the R package is described in Schuh and Furrer (2022). The R package contains three main functions:
strucDiv(): Scale-specific structural diversity quantification
The strucDiv() function quantifies structural diversity on a particular scale, which is defined by the size of the moving window. Within the area captured by the moving window, pixel values are considered as pairs at user defined distance and angle. A direction-invariant version is available. The frequencies of value pair occurrences are normalized by the total number of pixel pairs, which returns the gray level co-occurrence matrix (GLCM). The GLCM contains the empirical probabilities pi, j that pixel values vi and vj occur as a pair at the specified distance and angle(s), in the area captured by the moving window. Structural diversity is based on empirical probabilities of value co-occurrence. The output map is called a ‘(spatial) structural diversity map’ and it is returned as a raster layer. The output map represents structural diversity quantified on a specific scale. This method is described in detail in Schuh, Santos, de Jong, et al. (2022).
strucDivNest(): Multi-scale structural diversity quantification
The strucDivNest() function allows to combine information from different scales with an empirical Bayesian approach and a Beta-Binomial model. Two scales are nested inside each other - a larger, outer scale and a smaller, inner scale. Three different nesting schemes are available, whereby the inner scale is always a moving window. The outer scale can either be another mowing window, a block, or the domain (i.e. the input raster). The outer scale is used as prior information, and the inner scale serves as likelihood to estimate posterior probabilities of pixel value co-occurrences. Structural diversity is quantified based on these posterior probabilities. The final map is called a ‘(spatial) structural diversity map’ and is returned as a raster layer. The output map represents structural diversity, quantified across different spatial scales, which are defined by the outer scale and the inner scale. This method is described in detail in Schuh, Santos, Schaepman, et al. (2022).
strucDivDom(): Structural diversity of the domain
The strucDivDom() function calculates structural diversity of the domain, i.e. the input rater. The function returns the structural diversity values of the domain, and also the GLCM and the `diversity matrix’, which is described in detail Schuh and Furrer (2022).
Structural diversity metrics
Structural diversity metrics include: Structural diversity entropy, contrast, dissimilarity, homogeneity, and normalized entropy. Structural diversity entropy includes a difference weight δ ∈ {0, 1, 2}, which weighs the differences between pixel values vi and vj, using either absolute, or square weights. When δ = 0, structural diversity entropy corresponds to Shannon entropy (Shannon 1948). Shannon entropy has a scale-specific maximum, however, when the strucDivNest function is used, this maximum may be violated, depending on the posterior probabilities of pixel value co-occurrences. The values of structural diversity entropy with δ ∈ {1, 2} are not restricted and depend on the values of the input raster. Normalized entropy is Shannon entropy normalized over maximum entropy. When the strucDiv function is used, normalized entropy ranges between 0 and 1. When the strucDivNest function is used, normalized entropy may be larger than 1, depending on the posterior probabilities of value co-occurrences. Dissimilarity naturally employs δ = 1, contrast employs δ = 2, and homogeneity employs δ = 2 in the denominator. The values of dissimilarity and contrast are not restricted and depend on the values of the input raster. Homogeneity measures the closeness of pi, j to the diagonal in the GLCM, and ranges between 0 and 1 when the strucDiv function is used. When the strucDivNest function is used, homogeneity may be larger than 1, depending on the posterior probabilities of value co-occurrences.
For entropy, normalized entropy, and homogeneity, the possible number of gray levels (GL) is restricted, i.e. high numbers of GL lead to structureless diversity maps.
Installation
The StrucDiv package can be installed from CRAN with:
install.packages("StrucDiv")
Example
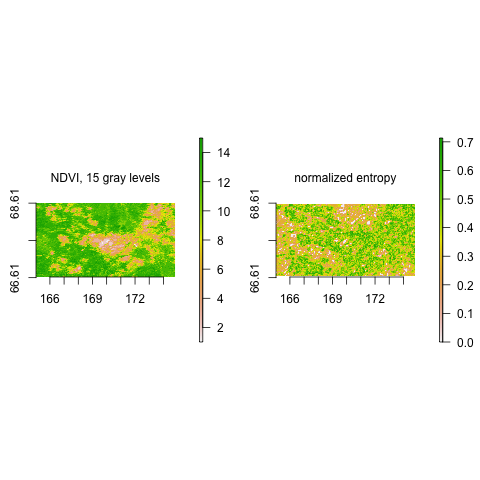
Calculate normalized entropy on Normalized Difference Vegetation Index (NDVI) data, which was binned to 15 gray levels (see data documentation). Scale is defined by window size length (WSL) 3, direct value neighbors are considered, and the direction-invariant version is employed.
ndvi.15gl <- raster(ndvi.15gl)
extent(ndvi.15gl) <- c(165.0205, 174.8301, 66.62804, 68.61332)
entNorm <- strucDiv(ndvi.15gl, wsl = 5, dist = 1, angle = "all", fun = entropyNorm,
na.handling = na.pass)
References
Haralick, R. M., K. Shanmugam, and I. Dinstein. 1973. “Textural Features for Image Classification.” IEEE Transactions on Systems, Man, and Cybernetics SMC-3 (6): 610–21. https://doi.org/10.1109/TSMC.1973.4309314.
Schuh, L., and R. Furrer. 2022. “StrucDiv: An R Package to Quantify Scale-Specific and Multi-Scale Spatial Structural Diversity in Continuous Raster Data.” In preparation.
Schuh, L., M. J. Santos, R. de Jong, M. Schaepman, and R. Furrer. 2022. “Structural Diversity Entropy: A Unified Diversity Measure to Detect Latent Landscape Features.” Under review.
Schuh, L., M. J. Santos, M. Schaepman, and R. Furrer. 2022. “An Empirical Bayesian Approach to Quantify Multi-Scale Spatial Structural Diversity in Remote Sensing Data.” Under review.
Shannon, Claude E. 1948. “A Mathematical Theory of Communication.” The Bell System Technical Journal 27 (3): 379–423. https://doi.org/10.1002/j.1538-7305.1948.tb01338.x.