Integration Unit Tests for Pharmacoepidemiological Studies.
TestGenerator
Does my cohort picked the correct number patients? Am I calculating an intersection in the right way? Is that the expected value for treatment duration? It just takes one incorrect parameter to get incoherent results in a pharmacoepidemiological study, and it is very challenging to test calculations on huge and complex databases.
That is why TestGenerator is useful to push a small sample of patients to unit test a study on the OMOP-CDM. It includes tools to create a blank CDM with a complete vocabulary and check if the code is doing what we expect in very specific cases.
This package is based on the unit testing written for the Eramus MC Ranitidine Study.
Installation
To install TestGenerator:
# CRAN version
install.packages("TestGenerator")
Example
The user can provide an Excel file (link to sample) or a set of CSV files that represent tables of the OMOP-CDM, with a micro population of just 8 patients for testing purposes.
readPatients() will read either Excel or CSVs, and then saves the data in a JSON file. This is useful if the user wants to create more than one Unit Test Definitions. If the parameter outputPath is NULL The files are saved in the testthat/testCases folder of the package. Alterna
TestGenerator::readPatients(filePath = "~/pathto/testPatients.xlsx",
testName = "test",
outputPath = NULL,
cdmVersion = "5.3")
Alternatively, the user can use the functions readPatients.xl or readPatients.csv directly.
TestGenerator::readPatients.xl(filePath = "~/pathto/testPatients.xlsx",
testName = "test",
outputPath = NULL,
cdmVersion = "5.3")
TestGenerator::readPatients.csv(filePath = "~/pathto/csv/files",
testName = "test",
outputPath = NULL,
cdmVersion = "5.3",
reduceLargeIds = FALSE)
patientCDM() pushes one of those Unit Test Definitions into a blank CDM reference with a complete version of the vocabulary. If the pathJSON parameter is NULL, TestGenerator will look for the JSON test files in the testthat/testCases folder.
cdm <- TestGenerator::patientsCDM(pathJson = NULL,
testName = "test",
cdmVersion = "5.3")
Now the user has a CDM reference with a complete vocabulary and just 8 patients.
filePath <- system.file("extdata/icu_sample_population.xlsx",
package = "TestGenerator")
outputPath <- file.path(tempdir(), "test")
dir.create(outputPath)
TestGenerator::readPatients(filePath = filePath,
testName = "test",
outputPath = outputPath,
cdmVersion = "5.3")
#> ✔ Unit Test Definition Created Successfully: 'test'
cdm <- TestGenerator::patientsCDM(pathJson = outputPath,
testName = "test",
cdmVersion = "5.3")
#> Note: method with signature 'DBIConnection#Id' chosen for function 'dbExistsTable',
#> target signature 'duckdb_connection#Id'.
#> "duckdb_connection#ANY" would also be valid
#> ! cdm name not specified and could not be inferred from the cdm source table
#> ✔ Patients pushed to blank CDM successfully
cdm[["person"]] %>% glimpse()
#> Rows: ??
#> Columns: 18
#> Database: DuckDB v1.0.0 [root@Darwin 24.1.0:R 4.4.1//private/var/folders/wm/s6fjrtt53ld72z03p47nkdvr0000gn/T/RtmpXvHHUw/file18235526d6af4.duckdb]
#> $ person_id <int> 1, 2, 3, 4, 5, 6, 7, 8
#> $ gender_concept_id <int> 8532, 8507, 8532, 8507, 8532, 8507, 8532, …
#> $ year_of_birth <int> 1980, 1990, 2000, 1980, 1990, 2000, 1980, …
#> $ month_of_birth <int> NA, NA, NA, NA, NA, NA, NA, NA
#> $ day_of_birth <int> NA, NA, NA, NA, NA, NA, NA, NA
#> $ birth_datetime <dttm> NA, NA, NA, NA, NA, NA, NA, NA
#> $ race_concept_id <int> 0, 0, 0, 0, 0, 0, 0, 0
#> $ ethnicity_concept_id <int> 0, 0, 0, 0, 0, 0, 0, 0
#> $ location_id <int> 0, 0, 0, 0, 0, 0, 0, 0
#> $ provider_id <int> 0, 0, 0, 0, 0, 0, 0, 0
#> $ care_site_id <int> 0, 0, 0, 0, 0, 0, 0, 0
#> $ person_source_value <chr> "0", "0", "0", "0", "0", "0", "0", "0"
#> $ gender_source_value <chr> "M", "F", "M", "F", "M", "F", "M", "F"
#> $ gender_source_concept_id <int> NA, NA, NA, NA, NA, NA, NA, NA
#> $ race_source_value <chr> NA, NA, NA, NA, NA, NA, NA, NA
#> $ race_source_concept_id <int> NA, NA, NA, NA, NA, NA, NA, NA
#> $ ethnicity_source_value <chr> NA, NA, NA, NA, NA, NA, NA, NA
#> $ ethnicity_source_concept_id <int> NA, NA, NA, NA, NA, NA, NA, NA
The reference can be used to create a cohort and create unit tests.
test_cohorts <- system.file("extdata",
"test_cohorts",
package = "TestGenerator")
cohort_set <- CDMConnector::readCohortSet(test_cohorts)
cdm <- CDMConnector::generateCohortSet(cdm,
cohort_set,
name = "test_cohorts")
#> ℹ Generating 3 cohorts
#> ℹ Generating cohort (1/3) - diazepam✔ Generating cohort (1/3) - diazepam [351ms]
#> ℹ Generating cohort (2/3) - hospitalisation✔ Generating cohort (2/3) - hospitalisation [272ms]
#> ℹ Generating cohort (3/3) - icu_visit✔ Generating cohort (3/3) - icu_visit [132ms]
cohortAttrition <- CDMConnector::attrition(cdm[["test_cohorts"]])
excluded_records <- cohortAttrition %>%
pull(excluded_records) %>%
sum()
expect_equal(excluded_records, 0)
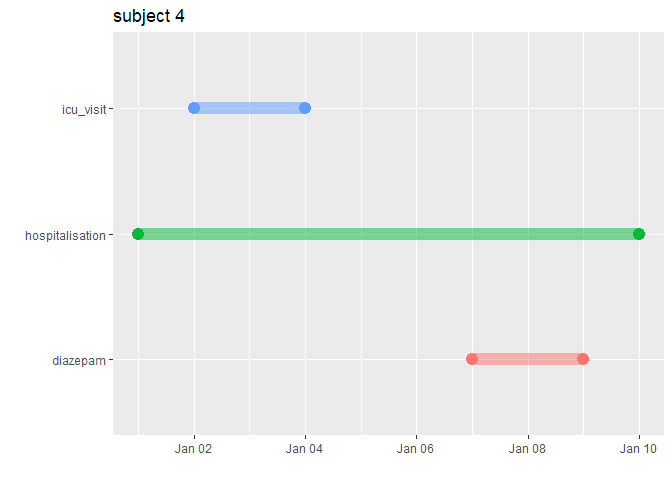
With graphCohort() it is possible to visualise the timeline for particular patient.
diazepam <- cdm[["test_cohorts"]] %>%
filter(cohort_definition_id == 1) %>%
collect()
hospitalisation <- cdm[["test_cohorts"]] %>%
filter(cohort_definition_id == 2) %>%
collect()
icu_visit <- cdm[["test_cohorts"]] %>%
filter(cohort_definition_id == 3) %>%
collect()
TestGenerator::graphCohort(subject_id = 4, list("diazepam" = diazepam,
"hospitalisation" = hospitalisation,
"icu_visit" = icu_visit))
#> Warning in geom_segment(aes(x = cohort_start_date, y = cohort, xend =
#> cohort_end_date, : Ignoring unknown aesthetics: fill