Description
Create View Tabs of Pipe Chains.
Description
Debugging pipe chains often consists of viewing the output after each step. This package adds RStudio addins and two functions that allow outputing each or select steps in a convenient way.
README.md
ViewPipeSteps 
Overview
ViewPipeSteps helps to debug pipe chains in a slightly more elegant fashion. Print/View debugging isn’t sexy, but instead of manually inserting %>% View() after each step, spice it up a bit by, e.g., highlighting the entire chain and calling the viewPipeChain addin:
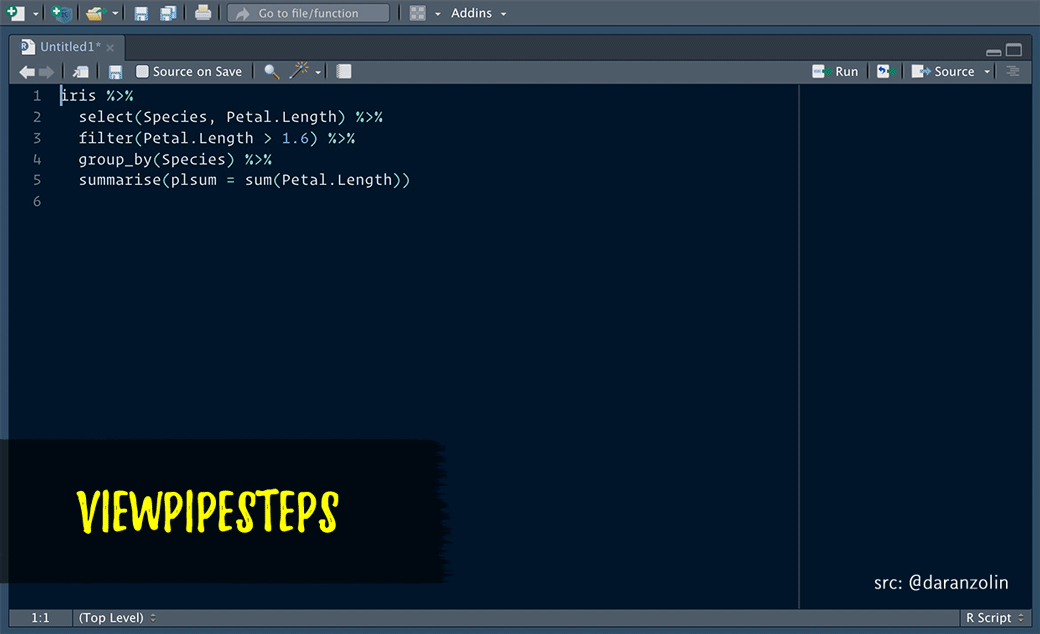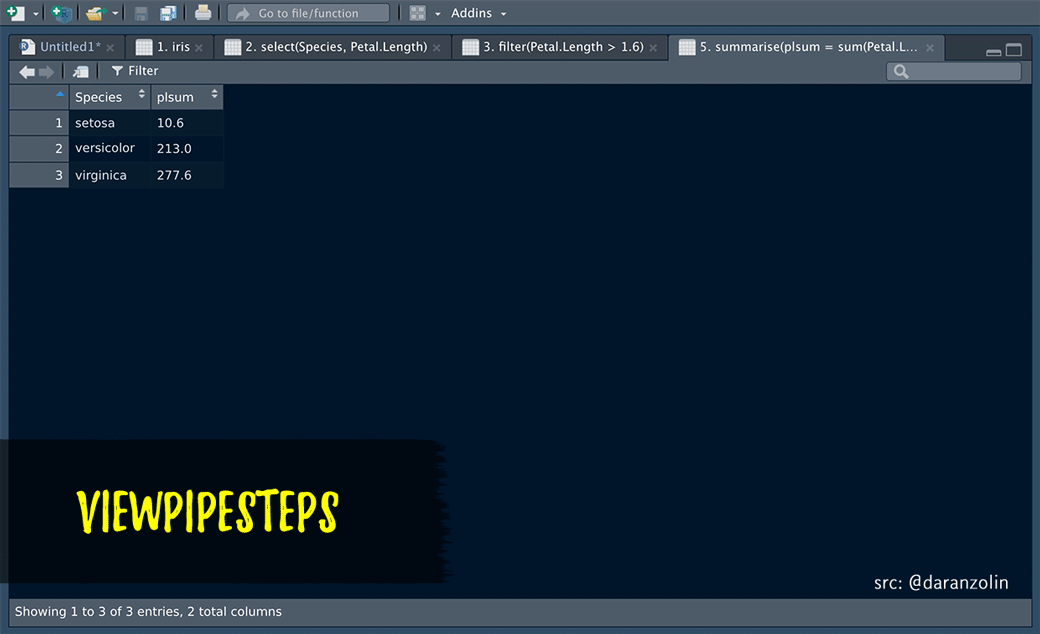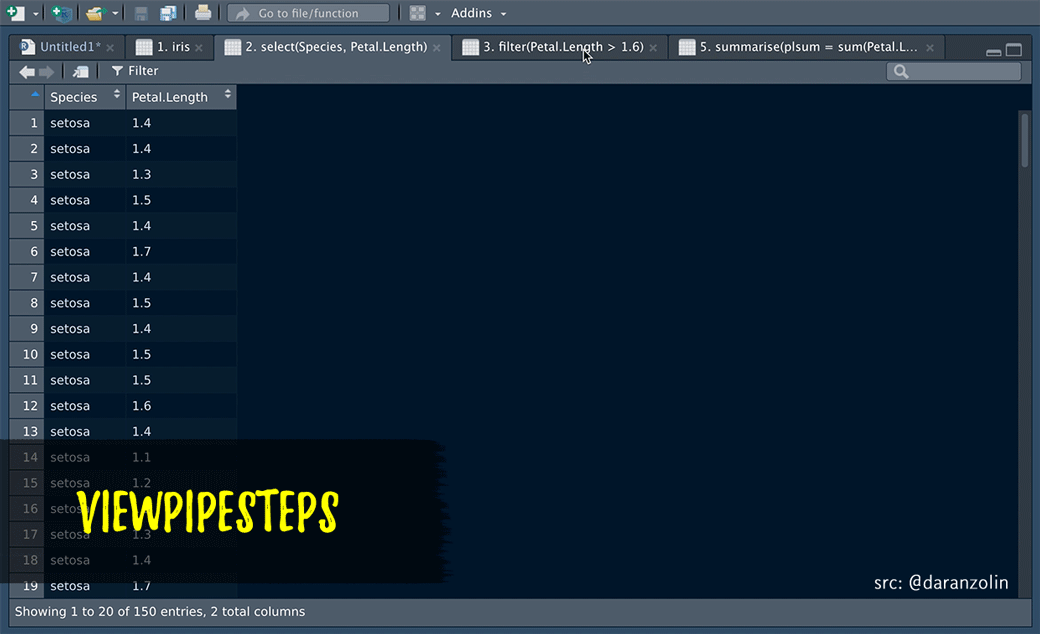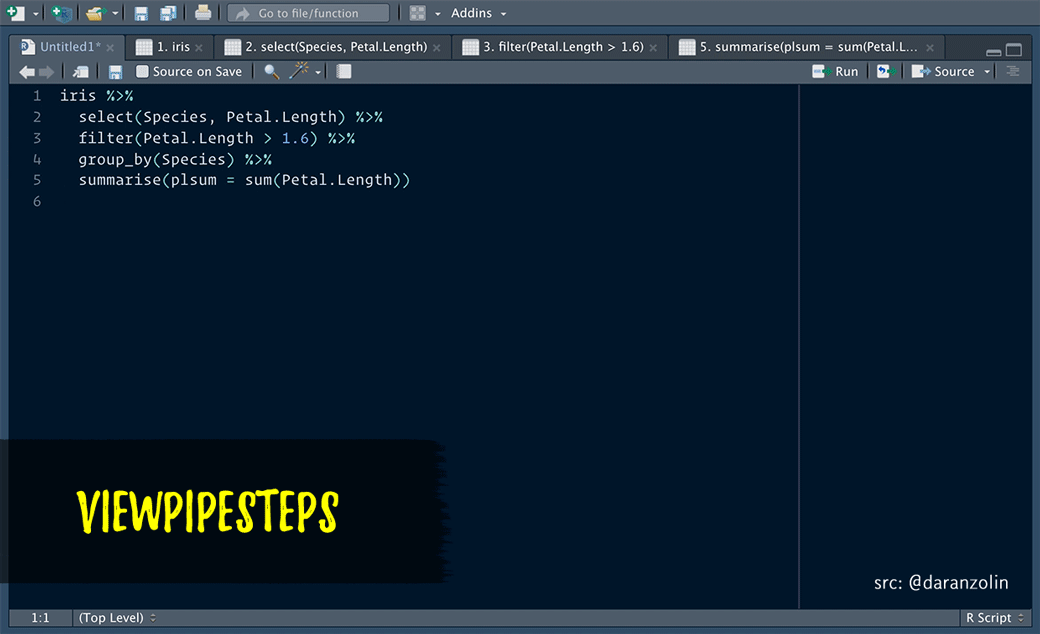
Thanks to @batpigandme for the the gif!
Alternatively, you can:
- Print each pipe step of the selction to the console by using the
printPipeChainaddin. - Print all pipe steps to the console by adding a print_pipe_steps() call to your pipe.
diamonds %>%
select(carat, cut, color, clarity, price) %>%
group_by(color) %>%
summarise(n = n(), price = mean(price)) %>%
arrange(desc(color)) %>%
print_pipe_steps() -> result
## 1. diamonds
## # A tibble: 53,940 x 10
## carat cut color clarity depth table price x y z
## <dbl> <ord> <ord> <ord> <dbl> <dbl> <int> <dbl> <dbl> <dbl>
## 1 0.23 Ideal E SI2 61.5 55 326 3.95 3.98 2.43
## 2 0.21 Premium E SI1 59.8 61 326 3.89 3.84 2.31
## 3 0.23 Good E VS1 56.9 65 327 4.05 4.07 2.31
## 4 0.290 Premium I VS2 62.4 58 334 4.2 4.23 2.63
## 5 0.31 Good J SI2 63.3 58 335 4.34 4.35 2.75
## 6 0.24 Very Good J VVS2 62.8 57 336 3.94 3.96 2.48
## 7 0.24 Very Good I VVS1 62.3 57 336 3.95 3.98 2.47
## 8 0.26 Very Good H SI1 61.9 55 337 4.07 4.11 2.53
## 9 0.22 Fair E VS2 65.1 61 337 3.87 3.78 2.49
## 10 0.23 Very Good H VS1 59.4 61 338 4 4.05 2.39
## # … with 53,930 more rows
## 2. select(carat, cut, color, clarity, price)
## # A tibble: 53,940 x 5
## carat cut color clarity price
## <dbl> <ord> <ord> <ord> <int>
## 1 0.23 Ideal E SI2 326
## 2 0.21 Premium E SI1 326
## 3 0.23 Good E VS1 327
## 4 0.290 Premium I VS2 334
## 5 0.31 Good J SI2 335
## 6 0.24 Very Good J VVS2 336
## 7 0.24 Very Good I VVS1 336
## 8 0.26 Very Good H SI1 337
## 9 0.22 Fair E VS2 337
## 10 0.23 Very Good H VS1 338
## # … with 53,930 more rows
## 4. summarise(n = n(), price = mean(price))
## # A tibble: 7 x 3
## color n price
## <ord> <int> <dbl>
## 1 D 6775 3170.
## 2 E 9797 3077.
## 3 F 9542 3725.
## 4 G 11292 3999.
## 5 H 8304 4487.
## 6 I 5422 5092.
## 7 J 2808 5324.
## 5. arrange(desc(color))
## # A tibble: 7 x 3
## color n price
## <ord> <int> <dbl>
## 1 J 2808 5324.
## 2 I 5422 5092.
## 3 H 8304 4487.
## 4 G 11292 3999.
## 5 F 9542 3725.
## 6 E 9797 3077.
## 7 D 6775 3170.
- Try your luck with the experimental
%P>%pipe variant that prints the output of the pipe’s left hand side prior to piping it to the right hand side.
diamonds %>%
select(carat, cut, color, clarity, price) %>%
group_by(color) %>%
summarise(n = n(), price = mean(price)) %P>%
arrange(desc(color)) -> result
## Printing diamonds %>% select(carat, cut, color, clarity, price) %>% group_by(color) %>% summarise(n = n(), price = mean(price))
## # A tibble: 7 x 3
## color n price
## <ord> <int> <dbl>
## 1 D 6775 3170.
## 2 E 9797 3077.
## 3 F 9542 3725.
## 4 G 11292 3999.
## 5 H 8304 4487.
## 6 I 5422 5092.
## 7 J 2808 5324.
Installation
devtools::install_github("daranzolin/ViewPipeSteps")
More Examples
Check tools/test_cases.R for more elaborate examples.
Future Work
- Verify that %P>% is implemented in a useful way and does it what it is supposed to do.