Additive Profile Clustering Algorithms.
adproclus
This package is an implementation of the additive profile clustering (ADPROCLUS) method in R. It can be used to obtain overlapping clustering models for object-by-variable data matrices. It also contains the low dimensional ADPROCLUS method, which achieves a simultaneous dimension reduction when searching for overlapping clusters. This can be used when the object-by-variable data contains a very large number of variables.
Installation
You can install the latest version from CRAN:
install.packages("adproclus")
Or install the development version of ADPROCLUS from GitHub with:
# install.packages("devtools")
devtools::install_github("henry-heppe/adproclus")
Example
This is a basic example which shows you how to use the regular ADPROCLUS and the low dimensional ADPROCLUS:
library("adproclus")
# import data
our_data <- adproclus::CGdata
# perform ADPROCLUS to get an overlapping clustering model
model_full <- adproclus(data = our_data, nclusters = 2)
# perform low dimensional ADPROCLUS to get an overlapping clustering model in terms of a smaller number of variables
model_lowdim <- adproclus_low_dim(data = our_data, nclusters = 3, ncomponents = 2)
To select the number of clusters (and the number of components in the low dimensional case) the package provides two model selection functions.
library("adproclus")
# estimate multiple ADPROCLUS models
models <- mselect_adproclus(data = CGdata, min_nclusters = 2, max_nclusters = 4)
# estimate multiple low dimensional ADPROCLUS models
models_lowdim <- mselect_adproclus_low_dim(data = CGdata, min_nclusters = 2, max_nclusters = 4, min_ncomponents = 1, max_ncomponents = 3)
# visualize models as a scree plot
plot_scree_adpc(models)
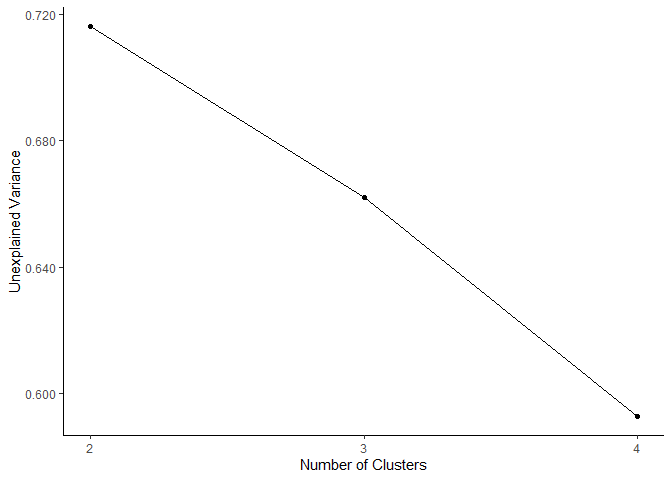
# visualize the low dimensional models as a scree plot
plot_scree_adpc(models_lowdim)
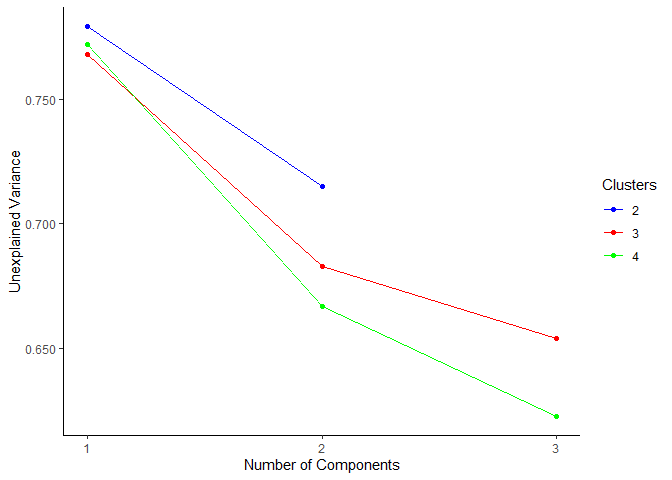
# select the best full dimensional model
best_model <- select_by_CHull(models)
# select a the conditionally optimal low dimensional model for each number of clusters
best_models_lowdim <- select_by_CHull(models_lowdim)
# visualize the preselected set of low dimensional models
plot_scree_adpc_preselected(best_models_lowdim)
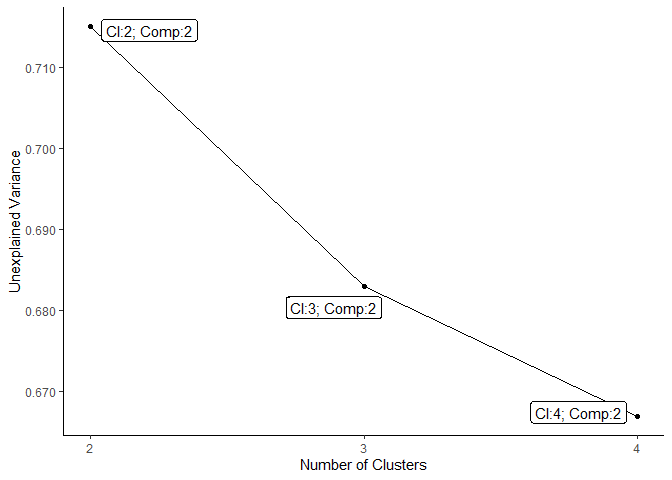
The package also provides functionality to obtain membership matrices, which the algorithm can start the alternating least squares procedure on. There are three different possibilities to obtain such matrices: random, semi-random and rational (see respective function documentation for details).
library("adproclus")
# import data
our_data <- adproclus::CGdata
# Obtaining a membership matrix were the entries are randomly assigned values of 0 or 1
start_allocation1 <- get_random(our_data, 3)
# Obtaining a membership matrix based on a profile matrix consisting of randomly selected rows of the data
start_allocation2 <- get_semirandom(our_data, 3)
# Obtaining a user-defined rational start profile matrix (here the first 3 rows of the data)
start_allocation3 <- get_rational(our_data, our_data[1:3, ])$A