Estimate Invasive and Alien Species (IAS) Introduction Rates.
alien: an R package for estimating alien introduction rates
alien is a package dedicated to easily estimate the introduction rates of alien species given first records data. It specializes in addressing the role of sampling on the pattern of discoveries, thus providing better estimates than using Generalized Linear Models which assume perfect immediate detection of newly introduced species.
Installation
You can install the CRAN version of the package with:
install.packages("alien")
You can install the development version of alien from GitHub with:
# install.packages("devtools")
devtools::install_github("hezibu/alien")
Basic Usage
For the most basic demonstration, let’s look at the data provided in Solow and Costello (2004) which describes discoveries of introduced species in the San Francisco estuary (California, USA) between the years 1850–1995. We’ll plot it in a cumulative form, replicating the plot from Solow and Costello (2004):
library(alien)
library(ggplot2)
data("sfestuary")
years <- seq_along(sfestuary) + 1850
ggplot()+
aes(x = years, y = cumsum(sfestuary))+
geom_line() +
coord_cartesian(ylim = c(0,150))+
scale_x_continuous(expand = c(0,0), breaks = seq(1860, 1980, 20)) +
scale_y_continuous(expand = c(0,0), breaks = seq(0, 150, 50)) +
ylab("Cumulative discoveries") + theme(axis.title.x = element_blank())
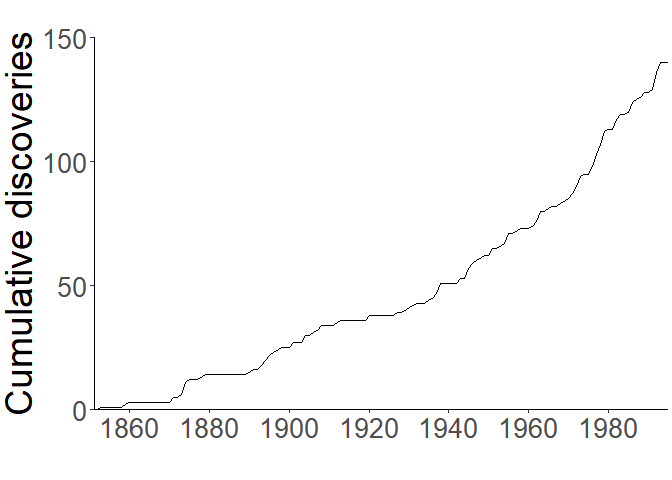
Model Fitting
As described thoroughly, these discoveries also entail trends in the probability of detecting new alien species. To estimate the introduction rate, ${\beta_1}$, from these data, we will fit the Solow and Costello model using the snc function. We can use the control argument to pass a list of options to optim which does the Maximum-Likelihood Estimation[^1]:
model <- snc(y = sfestuary, control = list(maxit = 1e4))
#> ! no data supplied, using time as independent variable
When only a vector describing discoveries is supplied, snc warns users that it uses the time as the independent variable, similar to the original S&C model.
The result is a list containing several objects:
names(model)
#> [1] "records" "convergence" "log-likelihood" "coefficients"
#> [5] "type" "fitted.values" "predict"
We’ll go over each.
Records
Shows the supplied records data.
model$records
#> [1] 0 0 1 0 0 0 0 0 1 1 0 0 0 0 0 0 0 0 0 0 2 0 1 5 1 0 0 1 1 0 0 0 0 0 0 0 0
#> [38] 0 0 1 1 0 2 2 2 1 1 1 0 0 2 0 0 3 0 1 1 2 0 0 0 1 1 0 0 0 0 0 0 2 0 0 0 0
#> [75] 0 0 1 0 1 1 1 1 0 0 1 1 2 4 0 0 0 0 2 0 4 2 1 1 1 0 3 0 1 1 4 0 1 1 0 0 1
#> [112] 2 4 0 1 1 0 1 1 1 2 3 4 1 0 3 5 4 5 1 0 4 2 0 1 4 1 1 2 0 1 7 4 0 0
Convergence
Did the optimation algorithm converge? This prints out the convergence code from optim:
model$convergence
#> [1] 0
| Code | Meaning/Troubleshooting |
|---|---|
| 0 | Successful convergence |
| 1 | Iteration limit maxit had been reached (increase maxit using control = list(maxit = number)) |
| 10 | Degeneracy of the Nelder-Mead simplex |
| 51 | Warning from the "L-BFGS-B"method; Use debug(snc) and check the optim component message for further details. |
| 52 | Error from the "L-BFGS-B"method; Use debug(snc) and check the optim component message for further details. |
log-likelihood
The log-likelihood at the end point of the algorithm (preferably at convergence). Can be used for model selection if needed:
model$`log-likelihood`
#> [1] 118.7776
coefficients
The parameter estimates.
beta0signifies ${\beta_0}$ - the intercept for ${\mu}$.gamma0signifies ${\gamma_0}$ - the intercept for ${\Pi}$.gamma2signifies ${\gamma_2}$ - and will only appear when thesncargumentgrowthis set toTRUE(the default).
model$coefficients
#> Estimate Std.Err
#> beta0 -1.12739745 1.835403
#> beta1 0.01401579 1.835403
#> gamma0 -185.89484996 15630.676343
#> gamma1 -79.80040427 7235.922667
#> gamma2 76.23985293 6339.859143
predict
The fitted ${\lambda_t}$ values of the model. The mean of the Poisson distribution from which the records are assumed to derive.
model$predict
#> mean lower_95 higher_95
#> 1 0.3284464 2.464968e-04 4.376407e+02
#> 2 0.3330822 6.848124e-06 1.620061e+04
#> 3 0.3377835 1.902532e-07 5.997150e+05
#> 4 0.3425512 5.285577e-09 2.220028e+07
#> 5 0.3473861 1.468428e-10 8.218113e+08
#> 6 0.3522893 4.079558e-12 3.042185e+10
#> 7 0.3572616 1.133375e-13 1.126158e+12
#> 8 0.3623042 3.148719e-15 4.168817e+13
#> 9 0.3674179 8.747707e-17 1.543215e+15
#> 10 0.3726038 2.430270e-18 5.712682e+16
#> 11 0.3778629 6.751728e-20 2.114724e+18
#> 12 0.3831963 1.875752e-21 7.828296e+19
#> 13 0.3886049 5.211175e-23 2.897883e+21
#> 14 0.3940898 1.447758e-24 1.072740e+23
#> 15 0.3996522 4.022133e-26 3.971074e+24
#> 16 0.4052931 1.117421e-27 1.470014e+26
#> 17 0.4110136 3.104396e-29 5.441708e+27
#> 18 0.4168148 8.624572e-31 2.014414e+29
#> 19 0.4226980 2.396061e-32 7.456970e+30
#> 20 0.4286641 6.656689e-34 2.760425e+32
#> 21 0.4347145 1.849348e-35 1.021856e+34
#> 22 0.4408502 5.137821e-37 3.782711e+35
#> 23 0.4470726 1.427379e-38 1.400286e+37
#> 24 0.4533828 3.965516e-40 5.183587e+38
#> 25 0.4597821 1.101692e-41 1.918863e+40
#> 26 0.4662716 3.060698e-43 7.103257e+41
#> 27 0.4728528 8.503170e-45 2.629488e+43
#> 28 0.4795269 2.362334e-46 9.733852e+44
#> 29 0.4862952 6.562988e-48 3.603283e+46
#> 30 0.4931590 1.823316e-49 1.333865e+48
#> 31 0.5001196 5.065500e-51 4.937709e+49
#> 32 0.5071786 1.407287e-52 1.827844e+51
#> 33 0.5143371 3.909697e-54 6.766323e+52
#> 34 0.5215967 1.086184e-55 2.504761e+54
#> 35 0.5289588 3.017615e-57 9.272138e+55
#> 36 0.5364248 8.383477e-59 3.432365e+57
#> 37 0.5439961 2.329081e-60 1.270595e+59
#> 38 0.5516743 6.470605e-62 4.703495e+60
#> 39 0.5594609 1.797651e-63 1.741142e+62
#> 40 0.5673574 4.994197e-65 6.445370e+63
#> 41 0.5753654 1.387478e-66 2.385951e+65
#> 42 0.5834864 3.854663e-68 8.832325e+66
#> 43 0.5917220 1.070895e-69 3.269555e+68
#> 44 0.6000738 2.975138e-71 1.210326e+70
#> 45 0.6085435 8.265468e-73 4.480390e+71
#> 46 0.6171328 2.296296e-74 1.658553e+73
#> 47 0.6258433 6.379523e-76 6.139641e+74
#> 48 0.6346767 1.772346e-77 2.272776e+76
#> 49 0.6436349 4.923897e-79 8.413374e+77
#> 50 0.6527194 1.367947e-80 3.114467e+79
#> 51 0.6619322 3.800403e-82 1.152915e+81
#> 52 0.6712751 1.055820e-83 4.267868e+82
#> 53 0.6807498 2.933259e-85 1.579882e+84
#> 54 0.6903582 8.149121e-87 5.848415e+85
#> 55 0.7001022 2.263973e-88 2.164969e+87
#> 56 0.7099838 6.289723e-90 8.014295e+88
#> 57 0.7200048 1.747398e-91 2.966736e+90
#> 58 0.7301673 4.854587e-93 1.098228e+92
#> 59 0.7404733 1.348692e-94 4.065427e+93
#> 60 0.7509246 3.746908e-96 1.504942e+95
#> 61 0.7615235 1.040958e-97 5.571002e+96
#> 62 0.7722721 2.891970e-99 2.062277e+98
#> 63 0.7831723 8.034412e-101 7.634147e+99
#> 64 0.7942263 2.232104e-102 2.826013e+101
#> 65 0.8054364 6.201187e-104 1.046135e+103
#> 66 0.8168047 1.722801e-105 3.872588e+104
#> 67 0.8283335 4.786252e-107 1.433557e+106
#> 68 0.8400250 1.329707e-108 5.306748e+107
#> 69 0.8518815 3.694165e-110 1.964455e+109
#> 70 0.8639054 1.026306e-111 7.272030e+110
#> 71 0.8760989 2.851262e-113 2.691964e+112
#> 72 0.8884646 7.921317e-115 9.965127e+113
#> 73 0.9010048 2.200685e-116 3.688896e+115
#> 74 0.9137220 6.113898e-118 1.365558e+117
#> 75 0.9266187 1.698551e-119 5.055029e+118
#> 76 0.9396975 4.718880e-121 1.871273e+120
#> 77 0.9529608 1.310990e-122 6.927089e+121
#> 78 0.9664113 3.642165e-124 2.564274e+123
#> 79 0.9800517 1.011859e-125 9.492443e+124
#> 80 0.9938846 2.811126e-127 3.513918e+126
#> 81 1.0079128 7.809815e-129 1.300784e+128
#> 82 1.0221389 2.169707e-130 4.815249e+129
#> 83 1.0365659 6.027837e-132 1.782512e+131
#> 84 1.0511965 1.674641e-133 6.598511e+132
#> 85 1.0660336 4.652455e-135 2.442640e+134
#> 86 1.0810801 1.292536e-136 9.042180e+135
#> 87 1.0963389 3.590897e-138 3.347239e+137
#> 88 1.1118132 9.976158e-140 1.239083e+139
#> 89 1.1275058 2.771556e-141 4.586843e+140
#> 90 1.1434200 7.699882e-143 1.697960e+142
#> 91 1.1595588 2.139166e-144 6.285518e+143
#> 92 1.1759253 5.942987e-146 2.326777e+145
#> 93 1.1925229 1.651069e-147 8.613275e+146
#> 94 1.2093547 4.586966e-149 3.188467e+148
#> 95 1.2264241 1.274342e-150 1.180308e+150
#> 96 1.2437344 3.540350e-152 4.369272e+151
#> 97 1.2612891 9.835731e-154 1.617419e+153
#> 98 1.2790915 2.732543e-155 5.987372e+154
#> 99 1.2971452 7.591496e-157 2.216409e+156
#> 100 1.3154538 2.109054e-158 8.204714e+157
#> 101 1.3340207 5.859332e-160 3.037225e+159
#> 102 1.3528497 1.627828e-161 1.124322e+161
#> 103 1.3719445 4.522399e-163 4.162021e+162
#> 104 1.3913087 1.256404e-164 1.540699e+164
#> 105 1.4109463 3.490516e-166 5.703368e+165
#> 106 1.4308611 9.697280e-168 2.111276e+167
#> 107 1.4510569 2.694079e-169 7.815533e+168
#> 108 1.4715378 7.484636e-171 2.893158e+170
#> 109 1.4923078 2.079367e-172 1.070991e+172
#> 110 1.5133710 5.776854e-174 3.964600e+173
#> 111 1.5347314 1.604914e-175 1.467618e+175
#> 112 1.5563933 4.458740e-177 5.432836e+176
#> 113 1.5783610 1.238718e-178 2.011130e+178
#> 114 1.6006387 3.441382e-180 7.444812e+179
#> 115 1.6232309 9.560779e-182 2.755925e+181
#> 116 1.6461419 2.656156e-183 1.020190e+183
#> 117 1.6693764 7.379280e-185 3.776544e+184
#> 118 1.6929387 2.050097e-186 1.398003e+186
#> 119 1.7168337 5.695538e-188 5.175135e+187
#> 120 1.7410659 1.582323e-189 1.915734e+189
#> 121 1.7656401 4.395978e-191 7.091676e+190
#> 122 1.7905612 1.221282e-192 2.625201e+192
#> 123 1.8158340 3.392940e-194 9.717982e+193
#> 124 1.8414635 9.426198e-196 3.597408e+195
#> 125 1.8674548 2.618767e-197 1.331690e+197
#> 126 1.8938130 7.275407e-199 4.929659e+198
#> 127 1.9205431 2.021239e-200 1.824864e+200
#> 128 1.9476506 5.615366e-202 6.755291e+201
#> 129 1.9751407 1.560050e-203 2.500677e+203
#> 130 2.0030187 4.334099e-205 9.257021e+204
#> 131 2.0312903 1.204091e-206 3.426769e+206
#> 132 2.0599609 3.345180e-208 1.268523e+208
#> 133 2.0890361 9.293513e-210 4.695826e+209
#> 134 2.1185218 2.581905e-211 1.738304e+211
#> 135 2.1484236 7.172997e-213 6.434861e+212
#> 136 2.1787475 1.992788e-214 2.382061e+214
#> 137 2.2094993 5.536322e-216 8.817925e+215
#> 138 2.2406853 1.538090e-217 3.264224e+217
#> 139 2.2723113 4.273091e-219 1.208352e+219
#> 140 2.3043838 1.187141e-220 4.473085e+220
#> 141 2.3369090 3.298093e-222 1.655849e+222
#> 142 2.3698932 9.162694e-224 6.129631e+223
#> 143 2.4033430 2.545561e-225 2.269070e+225
#> 144 2.4372649 7.072028e-227 8.399657e+226
#> 145 2.4716656 1.964736e-228 3.109390e+228
Plotting
Once we’ve fitted the model, we can use its fit to easily plot ${\lambda_t}$ along with the first records using the function plot_snc. Users can choose either annual or cumulative plots. Because the output is a ggplot object, it can easily be customized further:
plot_snc(model, cumulative = T) +
coord_cartesian(ylim = c(0,150))+
scale_y_continuous(expand = c(0,0), breaks = seq(0, 150, 50)) +
ylab("Cumulative discoveries") +
xlab("Years since first record in data")
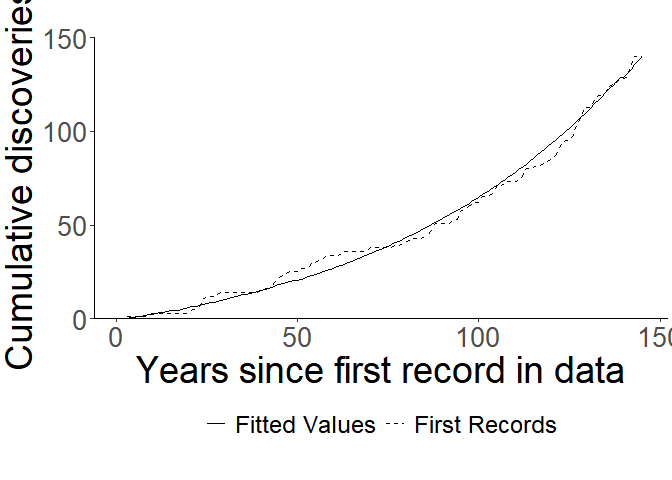
[^1]: In this case we increase maxiter so the algorithm will converge.