Likelihood Based Inference for ARIMA Modeling.
arima2
The goal of arima2 is to provide a set of tools to aid in the analysis of time series data in R. One such function is arima2::arima, which provides an interface to estimating Auto Regressive Integrated Moving Average (ARIMA) models using a random-restart algorithm. This function improves on the functionality of the stats::arima method, as it has the potential to increase the likelihood of the final output model. By design, the function cannot result in models with lower likelihoods than that of the stats::arima function. The potential for increased model likelihoods is obtained at the cost of computational efficiency. The function is approximately $n$ times slower than the stats::arima function, where $n$ is the number of random restarts. The benefit of trying multiple restarts becomes smaller as the number of available observations increases. Because the estimation of ARIMA models takes only a fraction of a second on relatively small data sets (less than a thousand observations), we are of the opinion that potential to increase model likelihoods is well worth this computational cost. The arima function implementation relies heavily on the source code of the stats::arima function.
Installation
# Install from CRAN
install.packages("arima2")
You can install the development version of arima2 from GitHub with:
# install.packages("devtools")
devtools::install_github("jeswheel/arima2")
Example
This is a basic example which shows you how to solve a common problem:
library(arima2)
#>
#> Attaching package: 'arima2'
#> The following object is masked from 'package:stats':
#>
#> arima
set.seed(83131)
# Get model coefficients from ARMA(2, 2)
coefs <- sample_ARMA_coef(order = c(2, 2))
# Get model intercept
intercept <- rnorm(1, sd = 50)
# Generate data from ARMA model
x <- intercept + arima.sim(
n = 100,
model = list(ar = coefs[grepl("^ar[[:digit:]]+", names(coefs))],
ma = coefs[grepl("^ma[[:digit:]]+", names(coefs))])
)
# Fit ARMA model using arima2 and stats::arima
arma2 <- arima(x, order = c(2, 0, 2), max_iters = 20)
arma <- stats::arima(x, order = c(2, 0, 2))
In the example above, the resulting log-likelihood of the stats::arima function is -142.24, and the log-likelihood of the arima function is -139.91. For this particular model and dataset, the random restart algorithm implemented in arima2 improved the model likelihood by 2.33 log-likelihood units.
Our package creates a new S3 object that we call Arima2, which extends the Arima class of the stats package. Once the model has been fit, our package includes some features that help diagnose the fitted model using this new child class. For example, ARMApolyroots function will return the AR or MA polynomial roots of the fitted model:
ARMApolyroots(arma2, type = 'AR')
#> [1] -4.464118+0i -15.209006+0i
ARMApolyroots(arma2, type = 'MA')
#> [1] -0.9870024+0.665084i -0.9870024-0.665084i
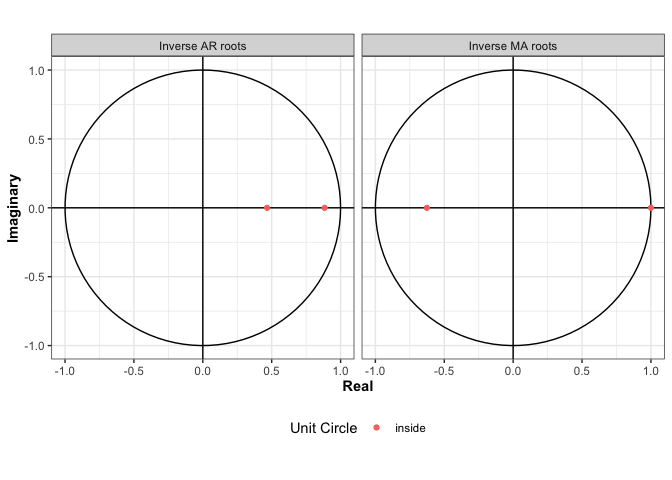
We have also implemented a plot.Arima2 function that uses the ggplot2 package so that we can visualize a fitted model. To compare the roots of the model fit using multiple restarts to the model fit using stats::arima, I will modify the class of the arma object so that it can easily be plotted.
class(arma) <- c("Arima2", class(arma))
plot(arma)
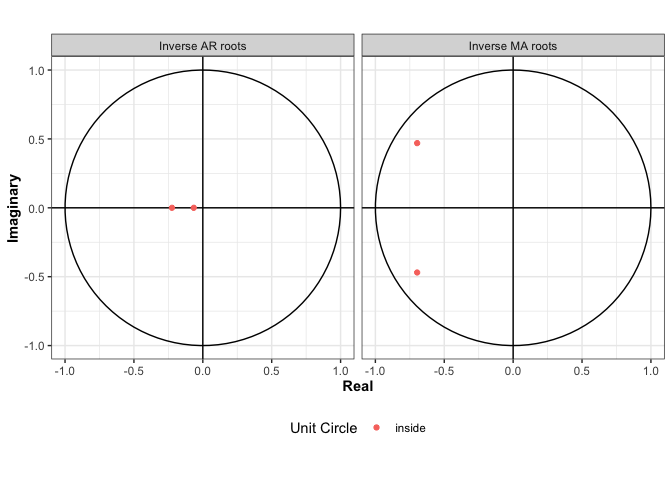
plot(arma2)
Finally, if a user would like help in determining an appropriate number of coefficients, we provide the aicTable function. The package also includes an aicTable function, which prints the AIC values for all ARMA$(p, d, q)$, with $p \leq P$, $q \leq Q$, and $d = D$:
set.seed(443252)
tab_results <- aicTable(x, P = 4, Q = 4, D = 0)
tab_results |> knitr::kable()
| MA0 | MA1 | MA2 | MA3 | MA4 | |
|---|---|---|---|---|---|
| AR0 | 368.7800 | 302.8397 | 290.2196 | 290.1671 | 291.4220 |
| AR1 | 319.4248 | 295.5320 | 289.8299 | 291.6313 | 292.9708 |
| AR2 | 303.4653 | 295.7739 | 291.8176 | 291.5108 | 293.0426 |
| AR3 | 296.6288 | 296.2828 | 292.9434 | 292.9987 | 293.5101 |
| AR4 | 295.1954 | 297.1895 | 294.9198 | 294.3391 | 294.9258 |
P <- which(tab_results == min(tab_results), arr.ind = TRUE)[1] - 1
Q <- which(tab_results == min(tab_results), arr.ind = TRUE)[2] - 1
print(paste0("p = ", P, "; q = ", Q))
#> [1] "p = 1; q = 2"
For more details about this package, please see our arXiv paper: arXiv:2310.01198