Automatic Cognostic Summaries.
autocogs
Cognostics are univariate statistics (or metrics) for a subset of data. When paired with the underlying data of visualizations, cognostics are a powerful tool for ordering and filtering the visualizations. add_panel_cogs() will automatically append cognostics for each plot player in a given panel column. The newly appended data can be fed into a trelliscopejs widget for easy viewing.
Installation
You can install autocogs from github with:
pak::pak("schloerke/autocogs")
Examples
Gapminder
library(autocogs)
library(tidyverse)
#> ── Attaching packages ─────────────────────────────────────── tidyverse 1.3.0 ──
#> ✔ ggplot2 3.3.3.9000 ✔ purrr 0.3.4
#> ✔ tibble 3.1.1 ✔ dplyr 1.0.5
#> ✔ tidyr 1.1.3 ✔ stringr 1.4.0
#> ✔ readr 1.4.0 ✔ forcats 0.5.1
#> ── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
#> ✖ dplyr::filter() masks stats::filter()
#> ✖ dplyr::lag() masks stats::lag()
library(gapminder)
# remotes::install_github("hafen/trelliscopejs")
# remotes::install_github("schloerke/trelliscopejs@autocogs")
library(trelliscopejs)
# Explore
p <-
ggplot(gapminder, aes(year, lifeExp)) +
geom_line(aes(group = country)) +
geom_smooth(method = "lm", formula = y ~ x)
p
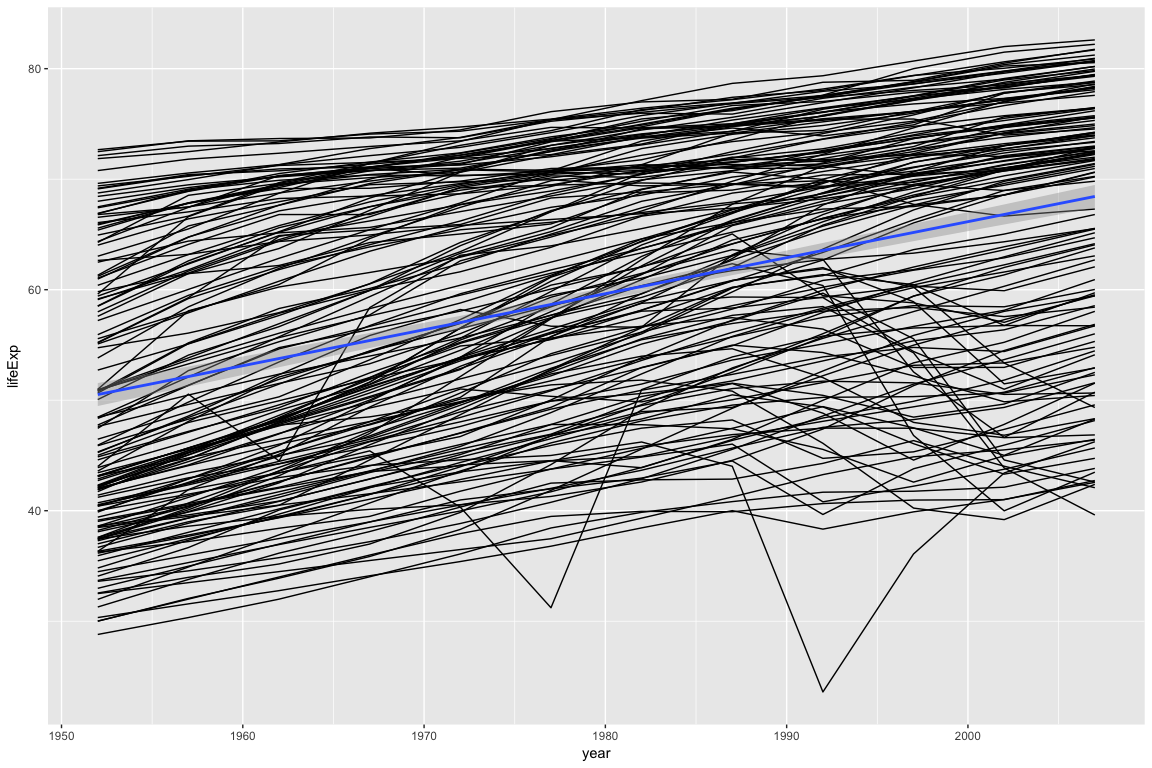
Looking at the plot above, most countries follow a linear trend: As the year increases, life expectancy goes up. A few countries do not follow a linear trend.
In the examples below, we will extract cognostics to aid in exploring the countries whose life expectancy is not linear.
trelliscopejs::facet_trelliscope()
ggplot(gapminder, aes(year, lifeExp)) +
geom_smooth(method = "lm", formula = y ~ x) +
geom_line() +
trelliscopejs::facet_trelliscope(
~ country + continent,
nrow = 3, ncol = 6,
self_contained = TRUE,
auto_cog = TRUE,
state = list(
# set the state to display the country, continent, and R^2 value
# sorted by ascending R^2 value
sort = list(trelliscopejs::sort_spec("_lm_r2")),
labels = c("country", "continent", "_lm_r2")
),
path = "readme-figs/facet"
)
#> using data from the first layer
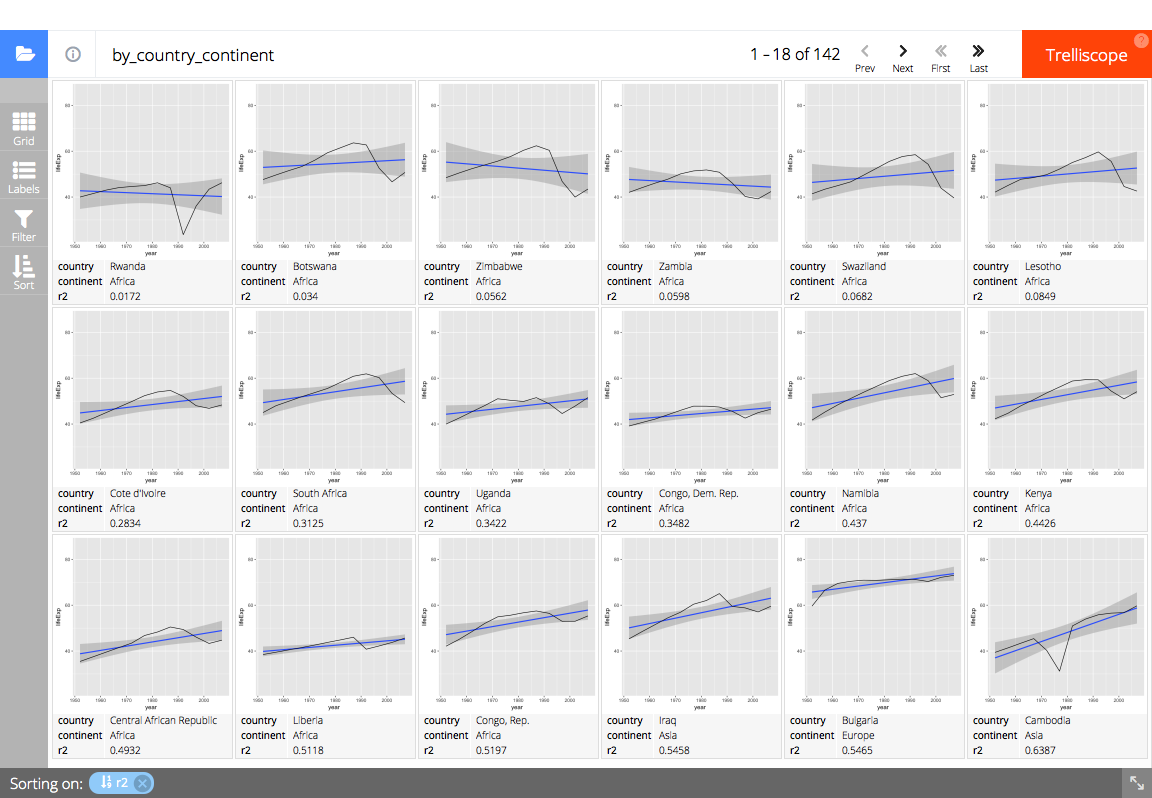
# (screen shot of trelliscopejs widget)
trelliscopejs::trelliscope()
This is a full, start to finish example how automatic cognostics could be inserted into a data exploration workflow.
# Find a consistent y range
y_range <- range(gapminder$lifeExp)
## # Set up data and panel column
gapminder %>%
group_by(country, continent) %>%
# nest the data according to the country and continent
nest() %>%
mutate(
# create a column of plots with a
# * line
# * linear model
panel = lapply(data, function(dt) {
ggplot(dt, aes(year, lifeExp)) +
geom_smooth(method = "lm", formula = y ~ x) +
geom_line() +
ylim(y_range[1], y_range[2])
})
) %>%
print() ->
gap_data
#> # A tibble: 142 x 4
#> # Groups: country, continent [142]
#> country continent data panel
#> <fct> <fct> <list> <list>
#> 1 Afghanistan Asia <tibble[,4] [12 × 4]> <gg>
#> 2 Albania Europe <tibble[,4] [12 × 4]> <gg>
#> 3 Algeria Africa <tibble[,4] [12 × 4]> <gg>
#> 4 Angola Africa <tibble[,4] [12 × 4]> <gg>
#> 5 Argentina Americas <tibble[,4] [12 × 4]> <gg>
#> 6 Australia Oceania <tibble[,4] [12 × 4]> <gg>
#> 7 Austria Europe <tibble[,4] [12 × 4]> <gg>
#> 8 Bahrain Asia <tibble[,4] [12 × 4]> <gg>
#> 9 Bangladesh Asia <tibble[,4] [12 × 4]> <gg>
#> 10 Belgium Europe <tibble[,4] [12 × 4]> <gg>
#> # … with 132 more rows
# Double check the plot worked...
# Look at the first panel (ggplot2 plot) of Afghanistan
gap_data$panel[[1]]
#!!!!!!!!!!
# Add cognostic information given the panel column plots
#!!!!!!!!!!
gap_data %>%
autocogs::add_panel_cogs() %>%
ungroup() %>%
# double check it was added
print(width = 100) ->
full_gap_data
#> # A tibble: 142 x 10
#> country continent data panel `_smooth`
#> <fct> <fct> <list> <list> <list>
#> 1 Afghanistan Asia <tibble[,4] [12 × 4]> <gg> <tibble[,3] [1 × 3]>
#> 2 Albania Europe <tibble[,4] [12 × 4]> <gg> <tibble[,3] [1 × 3]>
#> 3 Algeria Africa <tibble[,4] [12 × 4]> <gg> <tibble[,3] [1 × 3]>
#> 4 Angola Africa <tibble[,4] [12 × 4]> <gg> <tibble[,3] [1 × 3]>
#> 5 Argentina Americas <tibble[,4] [12 × 4]> <gg> <tibble[,3] [1 × 3]>
#> 6 Australia Oceania <tibble[,4] [12 × 4]> <gg> <tibble[,3] [1 × 3]>
#> 7 Austria Europe <tibble[,4] [12 × 4]> <gg> <tibble[,3] [1 × 3]>
#> 8 Bahrain Asia <tibble[,4] [12 × 4]> <gg> <tibble[,3] [1 × 3]>
#> 9 Bangladesh Asia <tibble[,4] [12 × 4]> <gg> <tibble[,3] [1 × 3]>
#> 10 Belgium Europe <tibble[,4] [12 × 4]> <gg> <tibble[,3] [1 × 3]>
#> `_lm` `_x` `_y` `_bivar` `_n`
#> <list> <list> <list> <list> <list>
#> 1 <tibble[,19] [1… <tibble[,5] [… <tibble[,5] [… <tibble[,2] [1… <tibble[,5] […
#> 2 <tibble[,19] [1… <tibble[,5] [… <tibble[,5] [… <tibble[,2] [1… <tibble[,5] […
#> 3 <tibble[,19] [1… <tibble[,5] [… <tibble[,5] [… <tibble[,2] [1… <tibble[,5] […
#> 4 <tibble[,19] [1… <tibble[,5] [… <tibble[,5] [… <tibble[,2] [1… <tibble[,5] […
#> 5 <tibble[,19] [1… <tibble[,5] [… <tibble[,5] [… <tibble[,2] [1… <tibble[,5] […
#> 6 <tibble[,19] [1… <tibble[,5] [… <tibble[,5] [… <tibble[,2] [1… <tibble[,5] […
#> 7 <tibble[,19] [1… <tibble[,5] [… <tibble[,5] [… <tibble[,2] [1… <tibble[,5] […
#> 8 <tibble[,19] [1… <tibble[,5] [… <tibble[,5] [… <tibble[,2] [1… <tibble[,5] […
#> 9 <tibble[,19] [1… <tibble[,5] [… <tibble[,5] [… <tibble[,2] [1… <tibble[,5] […
#> 10 <tibble[,19] [1… <tibble[,5] [… <tibble[,5] [… <tibble[,2] [1… <tibble[,5] […
#> # … with 132 more rows
# Display the panel and cognostics in a trelliscopejs widget
trelliscopejs::trelliscope(
full_gap_data, "gapminder life expectancy",
panel_col = "panel",
ncol = 6, nrow = 3,
auto_cog = FALSE,
self_contained = TRUE,
state = list(
# sort by ascending R^2 value (percent explained by linear model)
sort = list(trelliscopejs::sort_spec("_lm_r2")),
# display the country, continent, and R^2 value
labels = c("country", "continent", "_lm_r2")
),
path = "readme-figs/manually"
)
#> New names:
#> * min -> min...26
#> * max -> max...27
#> * mean -> mean...28
#> * median -> median...29
#> * var -> var...30
#> * ...
#> Warning: Removed 4 rows containing missing values (geom_smooth).
#> Warning: Removed 8 rows containing missing values (geom_smooth).
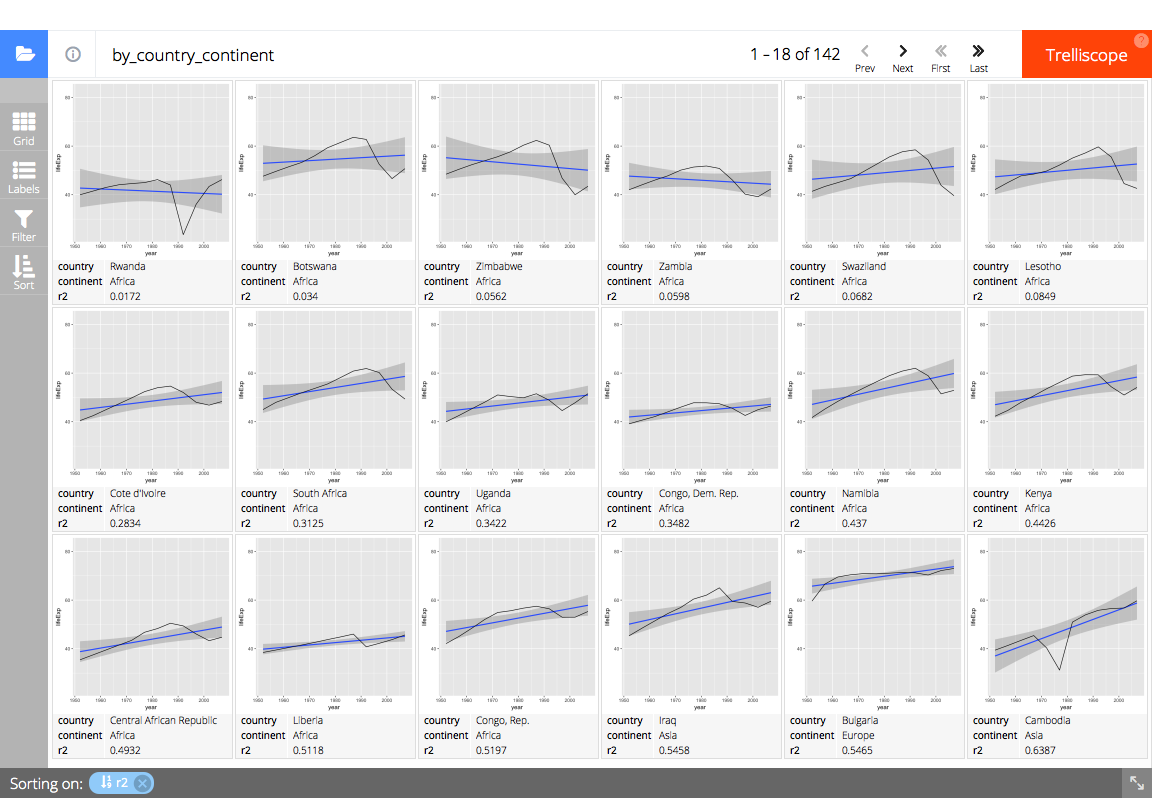
# (screen shot of trelliscopejs widget)
Custom Cognostics
add_cog_group()to add a custom cognostics group.add_layer_cogs()to call which cognostics groups should be executed for a given plot layer.
Using existing code from the autocogs package, we will add the univariate continuous cognostics group.
add_cog_group(
"univariate_continuous",
field_info("x", "continuous"),
"univariate metrics for continuous data",
function(x, ...) {
x_range <- range(x, na.rm = TRUE)
list(
min = cog_desc(x_range[1], "minimum of non NA data"),
max = cog_desc(x_range[2], "maximum of non NA data"),
mean = cog_desc(mean(x, na.rm = TRUE), "mean of non NA data"),
median = cog_desc(median(x, na.rm = TRUE), "median of non NA data"),
var = cog_desc(var(x, na.rm = TRUE), "variance of non NA data")
)
}
)
We can then call the 'univariate_continuous' cognostics group whenever a geom_rug layer is added in a ggplot2 plot object using the code below.
add_layer_cogs(
# load_all(); p <- qplot(x = 1, y = Sepal.Length, data = iris, geom = "boxplot"); plot_cogs(p)
"geom_boxplot",
"boxplot plot",
cog_group_df(
"univariate_continuous", "y", "_y",
"boxplot", "y", "_boxplot",
"univariate_counts", "y", "_n"
)
)
