Automating Choosing Statistical Tests.
automatedtest 
Automatically select and run the best statistical test for your data with just one line of code. Supporting one-sample-tests, two-sample-tests, multiple-sample-tests, and even correlations! automatedtests
What is automatedtest?
automatedtests is an R package designed to simplify statistical testing. It automatically analyzes your data, determines the most fitting statistical test (based on structure and content), and executes it. shortening the time spent deciding what test to use.
The package supports tidy data frames and a set of numeric/categorical vectors! non tidy data will have to be reshaped.
Features
- Auto-detects best statistical test based on your data type and structure.
- Handles tidy data: optional identifier exclusion.
- Returns an
AutomatedTestobject with many different results including the full test$get_result().
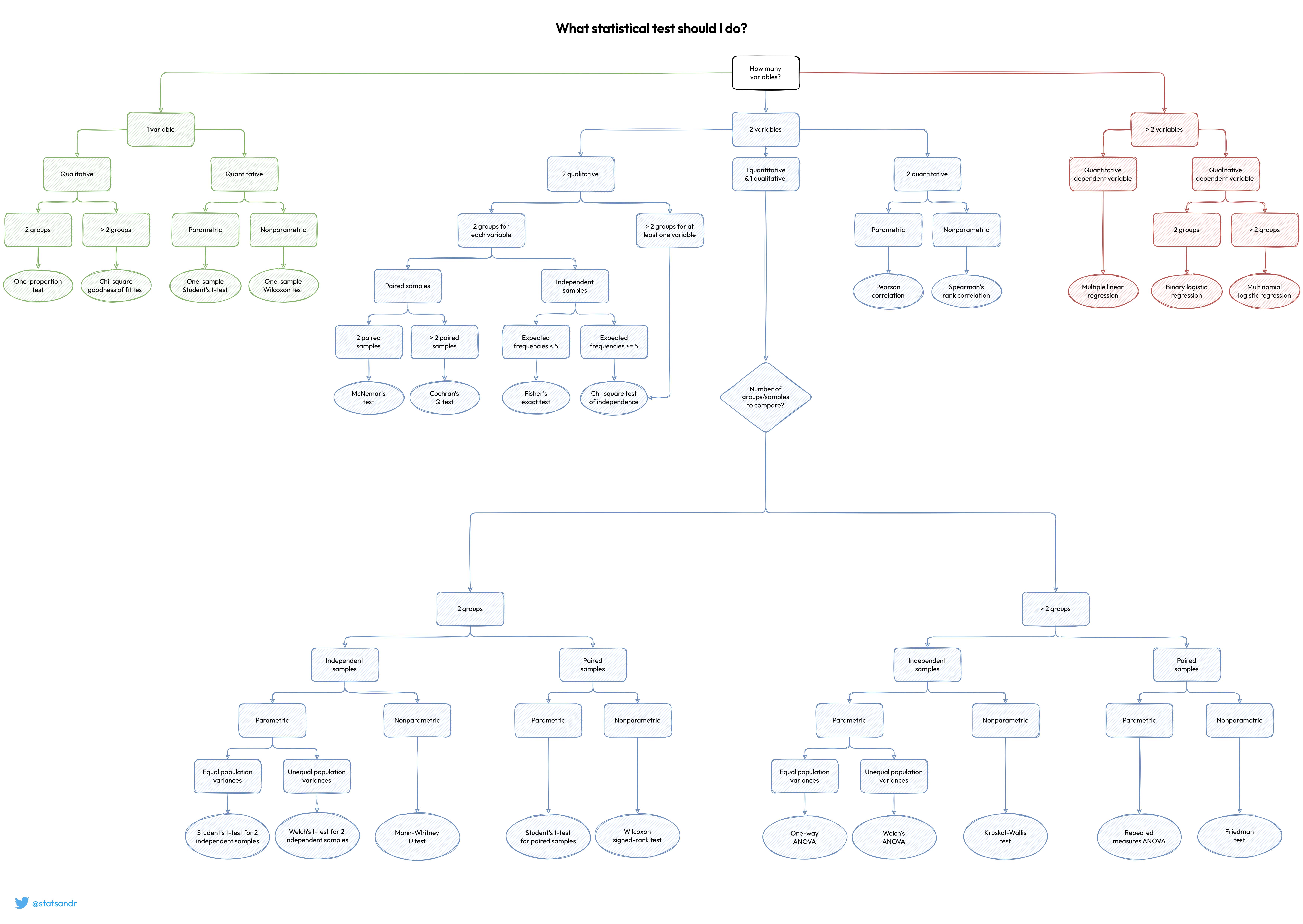
Supported Tests
| number | test |
|---|---|
| 1 | One-proportion test |
| 2 | Chi-square goodness-of-fit test |
| 3 | One-sample Student's t-test |
| 4 | One-sample Wilcoxon test |
| 5 | Multiple linear regression |
| 6 | Binary logistic regression |
| 7 | Multinomial logistic regression |
| 8 | Pearson correlation |
| 9 | Spearman's rank correlation |
| 10 | Cochran's Q test |
| 11 | McNemar's test |
| 12 | Fisher's exact test |
| 13 | Chi-square test of independence |
| 14 | Student's t-test for independent samples |
| 15 | Welch's t-test for independent samples |
| 16 | Mann-Whitney U test |
| 17 | Student's t-test for paired samples |
| 18 | Wilcoxon signed-rank test |
| 19 | One-way ANOVA |
| 20 | Welch's ANOVA |
| 21 | Repeated measures ANOVA |
| 22 | Kruskal-Wallis test |
| 23 | Friedman test |
Installation
You can install the package from CRAN:
install.packages("automatedtests")
# Load library
library(automatedtests)
Usage
Using a data frame
# Automatically runs appropriate test(s) on the cars dataset
test1 <- automatical_test(cars)
# Get quick overview
test1
# Get detailed results
test1$get_result()
Using individual vectors
# Compare Sepal.Length across Species
test2 <- automatical_test(iris$Species, iris$Sepal.Length)
test2$get_result()
One-sample tests
# Compare a numeric vector to a fixed value
automatical_test(c(3, 5, 4, 6, 7), compare_to = 5)
Arguments
| Argument | Description |
|---|---|
... | A data frame or multiple equal-length vectors |
compare_to | Value to compare against in one-sample tests (numeric or assumed uniform for categorical data) |
identifiers | Logical; if TRUE, the first column is treated as identifiers and excluded from testing |
paired | Logical; if TRUE, the test will become paired, by default FALSE |
Output
Returns an object of class AutomatedTest with methods and properties like:
print(object)- overview of executed test and its results.$get_result()- detailed summary of the test performed, containing all information including p.value, statistics etc.$get_test()- test type selected$is_parametric()- Whether the numeric feature were parametric$is_paired()- Returns if a paired test was used.$get_strength()- Shows the strength of the test/correlation. This is a different kind of value for each test. It will also return what the value is. These are the different types of data it can return:
coefficient – strength and direction of predictor effects
r – strength and direction of correlation
mean difference – size of difference between group means
statistic – test statistic indicating group difference or association
F statistic – variance ratio across group means
proportion – estimated proportion of successes in a sample
non-existent – no interpretable strength measure available
$get_parametric_list()- Returns a list of all numeric features' distributions and the parametric tests used.$get_datatypes()- Shows what type of data the features used in the corresponding test contain.$is_significant()- TRUE/FALSE if result is statistically significant (p.value < 0.05), to show the result in the blink of an eye!
Example Output
# Automated Test:
# Data: speed, dist
# Test: Spearman's rank correlation
# Test: Spearman's rank correlation
# Results:
# p.value: 8.824558e-14
# Strength: r = 0.83
# Significant: TRUE
Method to choose stastitical test
- By Antoine Soetewey
Dependencies
- R6
- MASS
- nnet
- nortest
- stats,
- DescTools
These are automatically handled during installation.
Author
Wouter Zeevat
License
This package is licensed under the GPL-3 License.
You can freely use, modify, and redistribute the software under the terms of the GNU General Public License v3 (GPL-3). The key conditions of the GPL-3 license are:
- You can use the package for personal, academic, or commercial purposes.
- If you modify the package and distribute it, you must distribute the source code of your modified version.
- Any derivative work must also be licensed under GPL-3.
For more information, see the full GPL-3 License.