Description
Goodman-Bacon Decomposition.
Description
Decomposition for differences-in-differences with variation in treatment timing from Goodman-Bacon (2018) <doi:10.3386/w25018>.
README.md
bacondecomp
bacondecomp is a package with tools to perform the Goodman-Bacon decomposition for differences-in-differences with variation in treatment timing. The decomposition can be done with and without time-varying covariates.
Installation
You can install bacondecomp 0.1.0 from CRAN:
install.packages("bacondecomp")
You can install the development version of bacondecomp from GitHub:
library(devtools)
install_github("evanjflack/bacondecomp")
Functions
bacon(): calculates all 2x2 differences-in-differences estimates and weights for the Bacon-Goodman decomposition.
Data
math_refom: Aggregated data from Goodman (2019, JOLE)castle: Data from Cheng and Hoekstra (2013, JHR)divorce:Data from Stevenson and Wolfers (2006, QJE)
Example
This is a basic example which shows you how to use the bacon() function to decompose the two-way fixed effects estimate of the effect of an education reform on future earnings following Goodman (2019, JOLE).
library(bacondecomp)
df_bacon <- bacon(incearn_ln ~ reform_math,
data = bacondecomp::math_reform,
id_var = "state",
time_var = "class")
#> type avg_est weight
#> 1 Earlier vs Later Treated 0.07117 0.06353
#> 2 Later vs Earlier Treated 0.04117 0.05265
#> 3 Treated vs Untreated 0.01211 0.88382
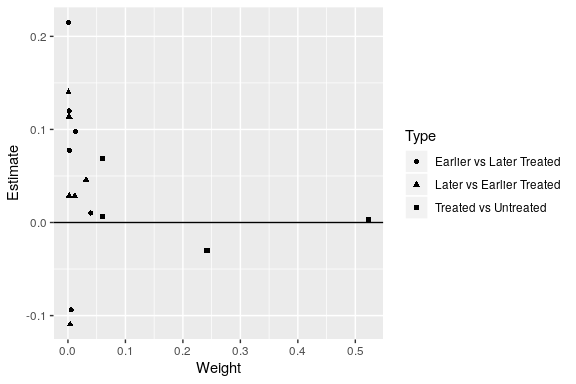
library(ggplot2)
ggplot(df_bacon) +
aes(x = weight, y = estimate, shape = factor(type)) +
geom_point() +
geom_hline(yintercept = 0) +
labs(x = "Weight", y = "Estimate", shape = "Type")
References
Goodman-Bacon, Andrew. 2018. “Difference-in-Differences with Variation in Treatment Timing.” National Bureau of Economic Research Working Paper Series No. 25018. doi: 10.3386/w25018.