Automated Standardized Assignment of the Banff Classification.
banffIT
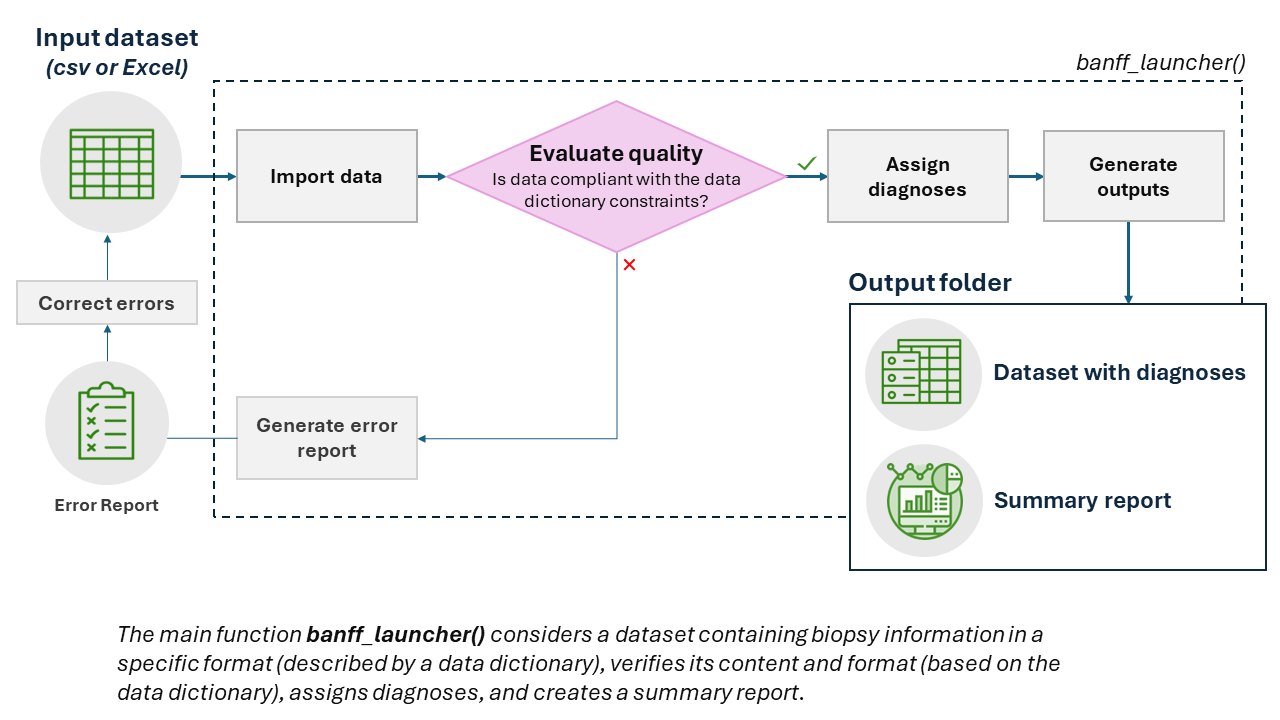
The banffIT package provides functions to assign standardized diagnoses using the Banff Classification (Category 1 to 6 diagnoses, including Acute and Chronic active T-cell mediated rejection as well as Active, Chronic active, and Chronic antibody mediated rejection). The main function banff_launcher()considers a minimal dataset containing biopsies information in a specific format (described by a data dictionary), verifies its content and format (based on the data dictionary), assigns diagnoses, and creates a summary report. It is possible to use different versions of the Banff classification.
Main functionality
DISCLAIMER
The banffIT package is distributed in the hope that it will be useful, but without any warranty. By using the banffIT package, you agree that its contents are used only for research purposes and not for making clinical decisions regarding patient care without consulting a pathologist. It is provided “as is” without warranty of any kind, either expressed or implied, including, but not limited to, the implied warranties of merchantability and fitness for a particular purpose.
In no event unless required by applicable law the author(s) will be liable to you for damages, including any general, special, incidental or consequential damages arising out of the use or inability to use the program (including but not limited to loss of data or data being rendered inaccurate or losses sustained by you or third parties or a failure of the program to operate with any other programs), even if the author(s) has been advised of the possibility of such damages.
Download section
also available in R using get_banff_dictionary()
also available in R using get_banff_template()
also available in R using get_banff_example()
Get started
Install the package and use the example file
# To install banffIT
install.packages('banffIT')
library(banffIT)
# If you need help with the package, please use:
banffIT_website()
# use example
version <- 2022
input_file <- system.file("extdata", paste0(version,"/banff_example.xlsx"),
package = "banffIT")
banff_launcher(
input_file = input_file,
output_folder = tempdir(), # 'folder_path/example'
version = version,
language = 'label:en',
option_filter = adequacy == 1,
detail = TRUE)