Description
Real-Time PCR Data Sets by Batsch et al. (2008).
Description
Real-time quantitative polymerase chain reaction (qPCR) data sets by Batsch et al. (2008) <doi:10.1186/1471-2105-9-95>. This package provides five data sets, one for each PCR target: (i) rat SLC6A14, (ii) human SLC22A13, (iii) pig EMT, (iv) chicken ETT, and (v) human GAPDH. Each data set comprises a five-point, four-fold dilution series. For each concentration there are three replicates. Each amplification curve is 45 cycles long. Original raw data file: <https://static-content.springer.com/esm/art%3A10.1186%2F1471-2105-9-95/MediaObjects/12859_2007_2080_MOESM5_ESM.xls>.
README.md
batsch
{batsch} provides real-time PCR data sets by Batsch et al. (2008) in tidy format. There are five data sets bundled with this package, one for each PCR target:
SLC6A14rfor rat SLC6A14SLC22A13hfor human SLC22A13EMTpfor pig EMTETTchfor chicken ETTGAPDHhfor human GAPDH
Each data set comprises a five-point, four-fold dilution series. For each concentration there are three replicates. Each amplification curve is 45 cycles long.
Installation
Install {batsch} from CRAN:
# Install from CRAN
install.packages("batsch")
You can instead install the development version of {batsch} from GitHub:
# install.packages("remotes")
remotes::install_github("ramiromagno/batsch")
Usage
Rat SLC6A14
library(batsch)
SLC6A14r |>
ggplot(mapping = aes(
x = cycle,
y = fluor,
group = interaction(dilution, replicate),
col = as.factor(dilution)
)) +
geom_line(linewidth = 0.5) +
labs(y = "Raw fluorescence", colour = "Dilution factor", title = "Rat SLC6A14") +
guides(color = guide_legend(override.aes = list(size = 0.5)))
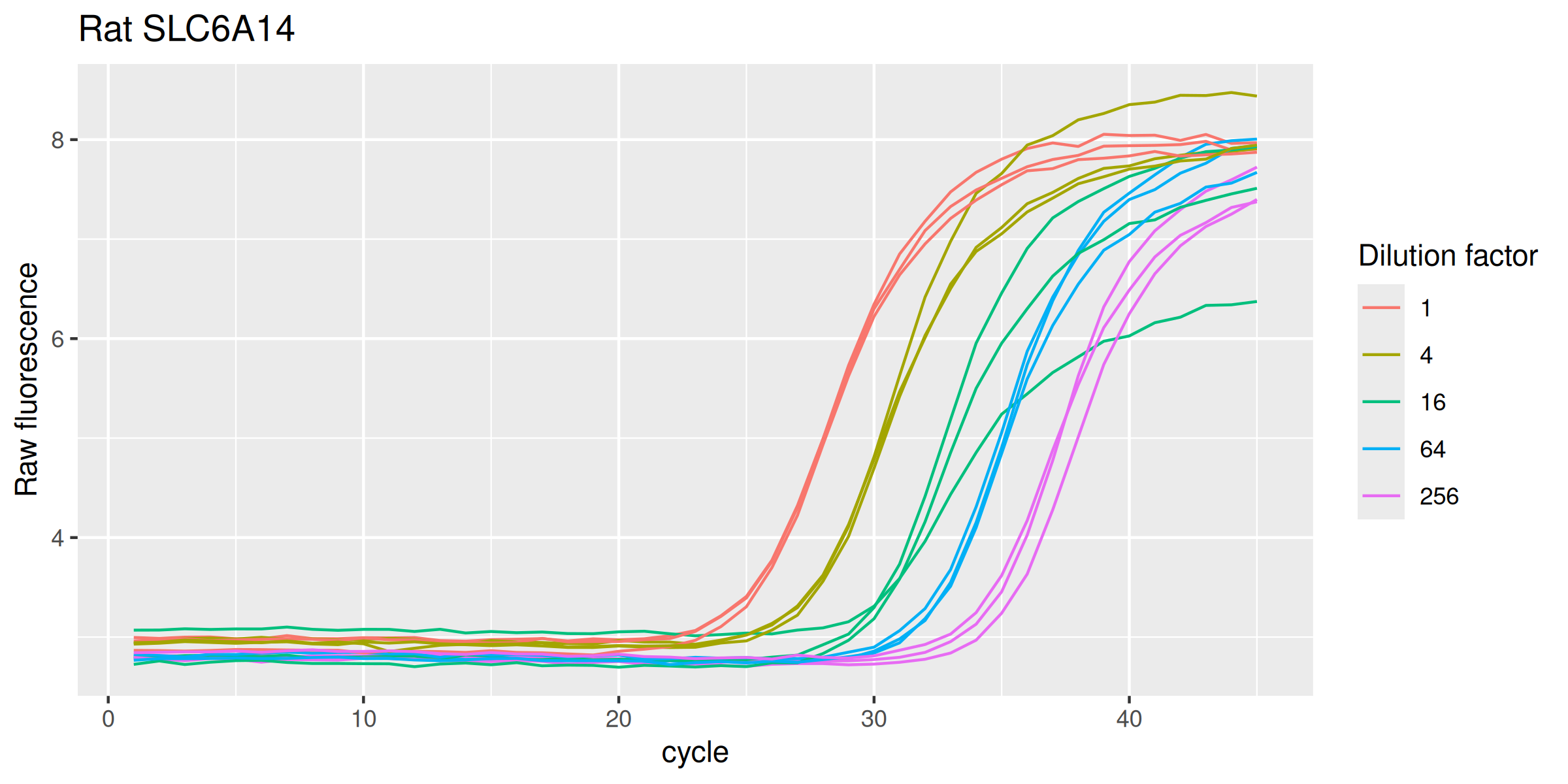
Human SLC22A13
SLC22A13h |>
ggplot(mapping = aes(
x = cycle,
y = fluor,
group = interaction(dilution, replicate),
col = as.factor(dilution)
)) +
geom_line(linewidth = 0.5) +
labs(y = "Raw fluorescence", colour = "Dilution factor", title = "Human SLC22A13") +
guides(color = guide_legend(override.aes = list(size = 0.5)))
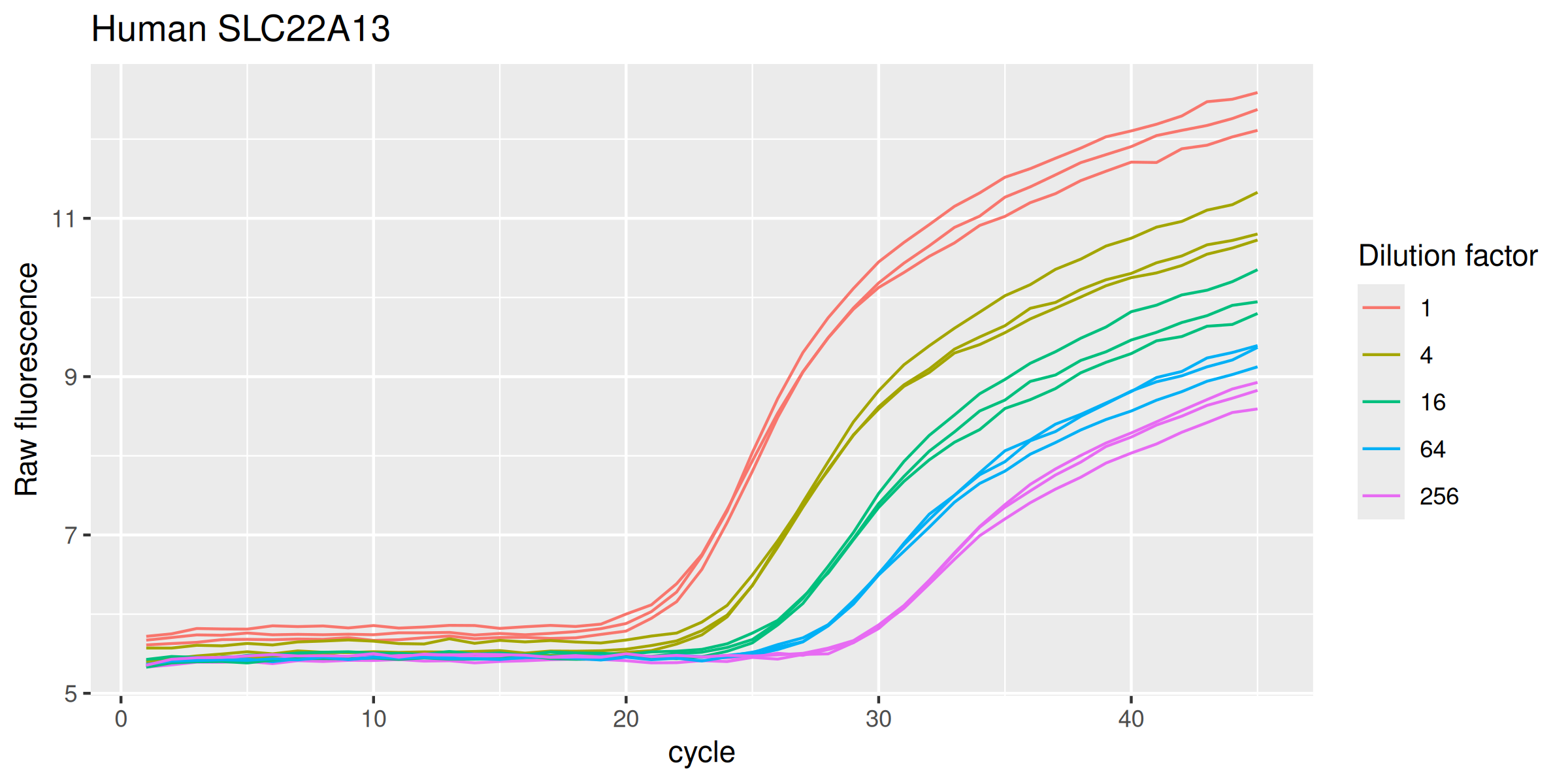
Pig EMT
EMTp |>
ggplot(mapping = aes(
x = cycle,
y = fluor,
group = interaction(dilution, replicate),
col = as.factor(dilution)
)) +
geom_line(linewidth = 0.5) +
labs(y = "Raw fluorescence", colour = "Dilution factor", title = "Pig EMT") +
guides(color = guide_legend(override.aes = list(size = 0.5)))
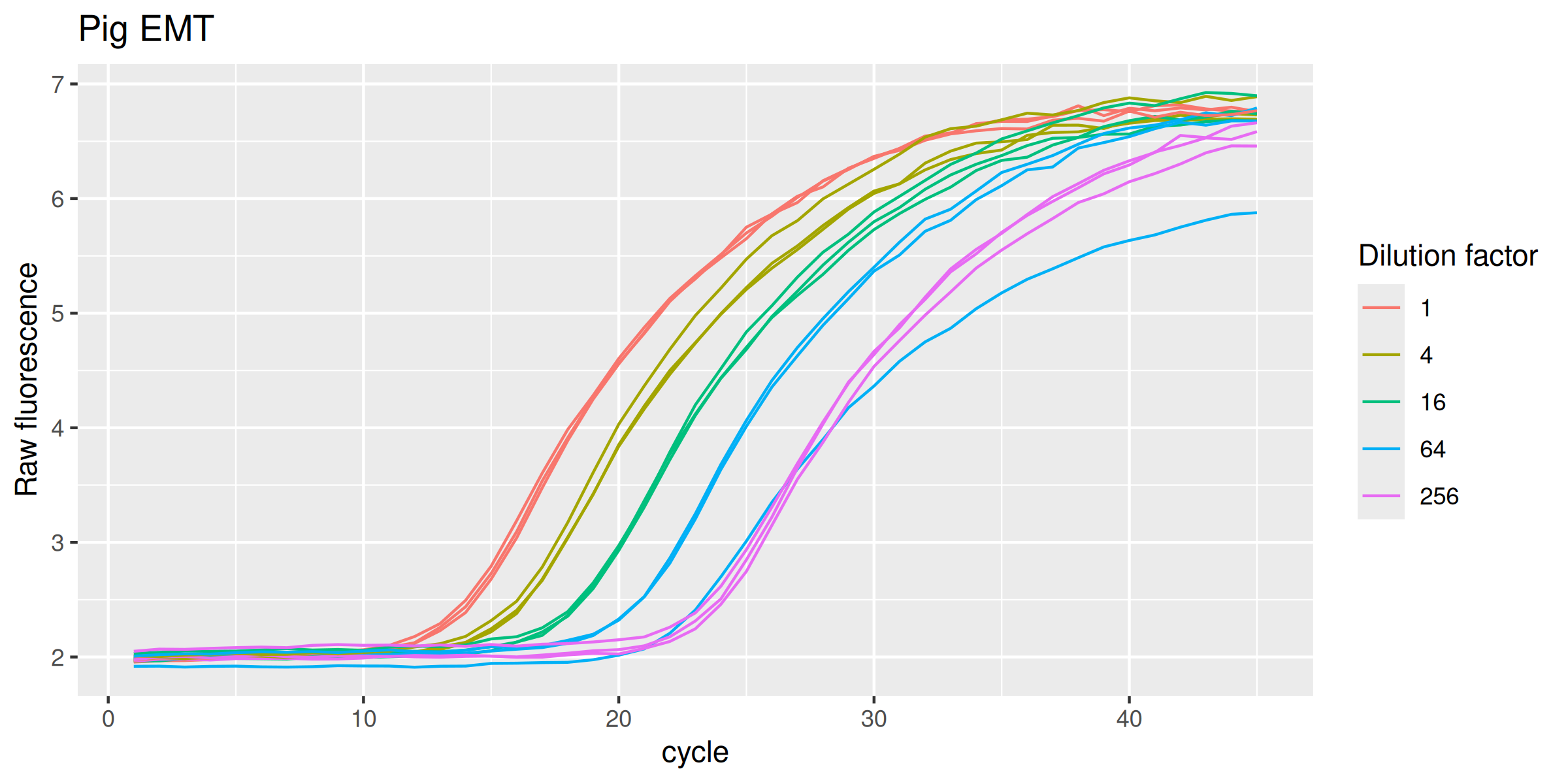
Chicken ETT
ETTch |>
ggplot(mapping = aes(
x = cycle,
y = fluor,
group = interaction(dilution, replicate),
col = as.factor(dilution)
)) +
geom_line(linewidth = 0.5) +
labs(y = "Raw fluorescence", colour = "Dilution factor", title = "Chicken ETT") +
guides(color = guide_legend(override.aes = list(size = 0.5)))
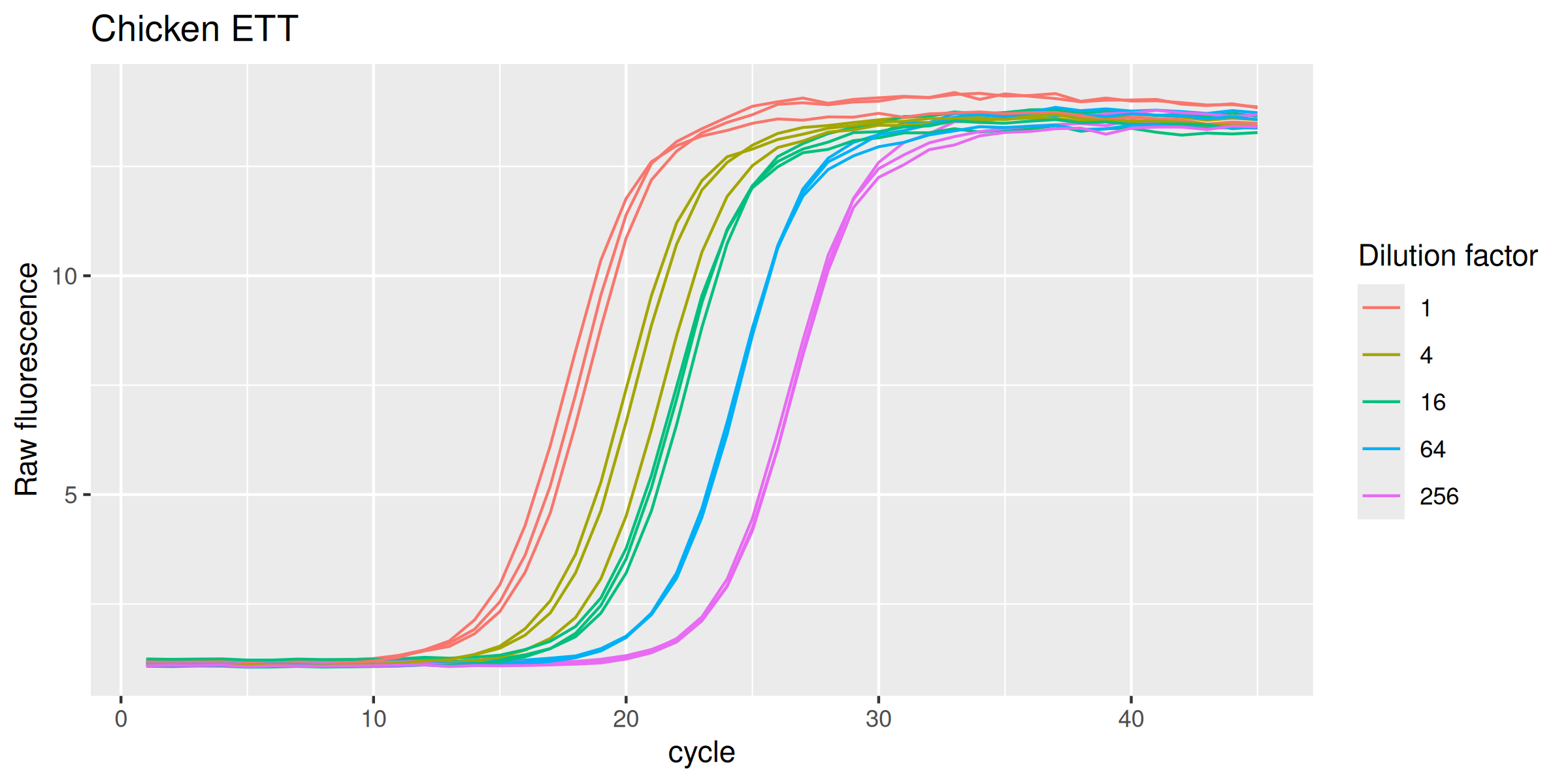
Human GAPDH
GAPDHh |>
ggplot(mapping = aes(
x = cycle,
y = fluor,
group = interaction(dilution, replicate),
col = as.factor(dilution)
)) +
geom_line(linewidth = 0.5) +
labs(y = "Raw fluorescence", colour = "Dilution factor", title = "Human GAPDH") +
guides(color = guide_legend(override.aes = list(size = 0.5)))

Code of Conduct
Please note that the {batsch} project is released with a Contributor Code of Conduct. By contributing to this project, you agree to abide by its terms.
References
- Anke Batsch, Andrea Noetel, Christian Fork, Anita Urban, Daliborka Lazic, Tina Lucas, Julia Pietsch, Andreas Lazar, Edgar Schömig & Dirk Gründemann. Simultaneous fitting of real-time PCR data with efficiency of amplification modeled as Gaussian function of target fluorescence. BMC Bioinformatics 9:95 (2008). doi: 10.1186/1471-2105-9-95.