Description
Mixed Effect Model with the Box-Cox Transformation.
Description
Inference on the marginal model of the mixed effect model with the Box-Cox transformation and on the model median differences between treatment groups for longitudinal randomized clinical trials. These statistical methods are proposed by Maruo et al. (2017) <doi:10.1002/sim.7279>.
README.md
bcmixed
The bcmixed package provides two categories of important functions: bcmarg and bcmmrm. The bcmarg function provides inferences on the marginal model of the mixed effect model with the Box-Cox transformation and the bcmmrm function provides inferences on the model median differences between treatment groups for longitudinal randomized clinical trials. These statistical methods are proposed by Maruo et al. (2017).
Installation
You can install the released version of bcmixed from CRAN with:
install.packages("bcmixed")
And the development version from GitHub with:
# install.packages("devtools")
devtools::install_github("kzkzmr/bcmixed")
Example
This is a basic example which shows you how to solve a common problem:
library(bcmixed)
data(aidscd4)
# Marginal model of mixed model with the Box-Cox transformation
res1 <- bcmarg(cd4 ~ as.factor(treatment) * as.factor(weekc) + age,
data = aidscd4, time = weekc, id = id)
summary(res1)
#> Box-Cox transformed mixed model analysis
#> Formula: cd4 ~ as.factor(treatment) * as.factor(weekc) + age
#> Time: weekc
#> ID: id
#> Covariance structure: "UN"
#> Data: aidscd4
#> Log-likelihood: -13623.31
#> Estimated transformation parameter: 0.183
#>
#> Coefficients on the transformed scale:
#> Value Std.Error t-value p-value
#> (Intercept) 3.1483 0.25917 12.147 0.000
#> as.factor(treatment)2 0.2967 0.16995 1.746 0.081
#> as.factor(treatment)3 0.4716 0.16946 2.783 0.005
#> as.factor(treatment)4 0.7614 0.16763 4.542 0.000
#> as.factor(weekc)16 -0.2254 0.09248 -2.437 0.015
#> as.factor(weekc)24 -0.4863 0.09781 -4.972 0.000
#> as.factor(weekc)32 -0.7246 0.10834 -6.688 0.000
#> age 0.0207 0.00612 3.384 0.001
#> as.factor(treatment)2:as.factor(weekc)16 -0.1386 0.12994 -1.067 0.286
#> as.factor(treatment)3:as.factor(weekc)16 -0.0500 0.12943 -0.387 0.699
#> as.factor(treatment)4:as.factor(weekc)16 0.0682 0.12896 0.529 0.597
#> as.factor(treatment)2:as.factor(weekc)24 -0.2078 0.13763 -1.510 0.131
#> as.factor(treatment)3:as.factor(weekc)24 -0.0235 0.13758 -0.171 0.864
#> as.factor(treatment)4:as.factor(weekc)24 0.0821 0.13754 0.597 0.551
#> as.factor(treatment)2:as.factor(weekc)32 -0.1034 0.15508 -0.667 0.505
#> as.factor(treatment)3:as.factor(weekc)32 -0.1018 0.15353 -0.663 0.507
#> as.factor(treatment)4:as.factor(weekc)32 0.1436 0.15162 0.947 0.344
#>
#> Covariance parameters on the transformed scale:
#> UN(1,1) UN(1,2) UN(1,3) UN(1,4) UN(2,2) UN(2,3) UN(2,4) UN(3,3) UN(3,4) UN(4,4)
#> 3.72 2.76 2.61 2.34 3.47 2.67 2.56 3.24 2.68 3.27
#>
#> Correlations on the transformed scale:
#> 8 16 24 32
#> 8 1.000 0.767 0.751 0.670
#> 16 0.767 1.000 0.797 0.761
#> 24 0.751 0.797 1.000 0.821
#> 32 0.670 0.761 0.821 1.000
# Box-Cox transformation for the baseline
lmd.bl <- bcmarg(cd4.bl ~ 1, data = aidscd4[aidscd4$weekc == 8, ])$lambda
aidscd4$cd4.bl.tr <- bct(aidscd4$cd4.bl, lmd.bl)
# Inference on model median differences between groups at each time point
res2 <- bcmmrm(outcome = cd4, group = treatment, data = aidscd4, time = weekc,
id = id, covv = c("cd4.bl.tr", "sex"), cfactor = c(0, 1),
glabel = c("Zid/Did", "Zid+Zal", "Zid+Did", "Zid+Did+Nev"))
# Summarize
print(res2)
#> Model median estimation based on MMRM with Box-Cox transformation
#> Outcome: cd4
#> Group: treatment
#> Time: weekc
#> ID: id
#> Covariate(s): c("cd4.bl.tr", "sex")
#> Covariance structure: "UN"
#> Data: aidscd4
#> Estimated transformation parameter: 0.154
#> Log-likelihood: -13322.36
#>
#> Model median estimates (row: group, col: time):
#> treatment | weekc 8 16 24 32
#> 1 Zid/Did 18.9 16.5 14.1 12.1
#> 2 Zid+Zal 22.0 17.9 14.6 13.5
#> 3 Zid+Did 24.5 20.9 18.2 15.1
#> 4 Zid+Did+Nev 30.1 27.8 24.2 21.8
summary(res2)
#> Model median inference based on MMRM with Box-Cox transformation
#>
#> Data and variable information:
#> Outcome: cd4
#> Group: treatment
#> Time: weekc
#> ID: id
#> Covariate(s): c("cd4.bl.tr", "sex")
#> Data: aidscd4
#>
#> Analysis information:
#> Covariance structure: "UN"
#> Robust inference: TRUE
#> Empirical small sample adjustment: TRUE
#> Confidence level: 0.95
#>
#> Analysis results:
#> Estimated transformation parameter: 0.154
#>
#>
#> Model median inferences for weekc = 8
#>
#> treatment median SE lower.CL upper.CL
#> 1 Zid/Did 18.9 0.862 17.2 20.6
#> 2 Zid+Zal 22.0 1.124 19.8 24.2
#> 3 Zid+Did 24.5 1.465 21.6 27.4
#> 4 Zid+Did+Nev 30.1 1.597 27.0 33.3
#>
#>
#> Model median inferences for weekc = 16
#>
#> treatment median SE lower.CL upper.CL
#> 1 Zid/Did 16.5 0.799 15.0 18.1
#> 2 Zid+Zal 17.9 0.932 16.1 19.8
#> 3 Zid+Did 20.9 1.207 18.5 23.2
#> 4 Zid+Did+Nev 27.8 1.596 24.6 30.9
#>
#>
#> Model median inferences for weekc = 24
#>
#> treatment median SE lower.CL upper.CL
#> 1 Zid/Did 14.1 0.716 12.7 15.5
#> 2 Zid+Zal 14.6 0.864 12.9 16.3
#> 3 Zid+Did 18.2 1.175 15.9 20.5
#> 4 Zid+Did+Nev 24.2 1.510 21.3 27.2
#>
#>
#> Model median inferences for weekc = 32
#>
#> treatment median SE lower.CL upper.CL
#> 1 Zid/Did 12.1 0.662 10.8 13.4
#> 2 Zid+Zal 13.5 0.813 11.9 15.1
#> 3 Zid+Did 15.1 1.019 13.1 17.1
#> 4 Zid+Did+Nev 21.8 1.376 19.1 24.5
#>
#>
#> Inferences of model median difference between groups ( treatment 1 - treatment 0 ) for weekc = 8
#>
#> treatment 1 treatment 0 delta SE lower.CL upper.CL t.value p.value
#> 1 Zid+Zal Zid/Did 3.12 1.40 0.363 5.87 2.22 0.027
#> 2 Zid+Did Zid/Did 5.64 1.69 2.325 8.96 3.34 0.001
#> 3 Zid+Did+Nev Zid/Did 11.25 1.80 7.711 14.80 6.24 0.000
#> 4 Zid+Did Zid+Zal 2.53 1.83 -1.059 6.12 1.39 0.167
#> 5 Zid+Did+Nev Zid+Zal 8.14 1.93 4.349 11.93 4.22 0.000
#> 6 Zid+Did+Nev Zid+Did 5.61 2.16 1.372 9.85 2.60 0.010
#>
#>
#> Inferences of model median difference between groups ( treatment 1 - treatment 0 ) for weekc = 16
#>
#> treatment 1 treatment 0 delta SE lower.CL upper.CL t.value p.value
#> 1 Zid+Zal Zid/Did 1.38 1.21 -1.0015 3.75 1.14 0.256
#> 2 Zid+Did Zid/Did 4.31 1.43 1.4940 7.12 3.01 0.003
#> 3 Zid+Did+Nev Zid/Did 11.22 1.78 7.7118 14.73 6.29 0.000
#> 4 Zid+Did Zid+Zal 2.93 1.50 -0.0162 5.88 1.95 0.051
#> 5 Zid+Did+Nev Zid+Zal 9.84 1.84 6.2318 13.46 5.36 0.000
#> 6 Zid+Did+Nev Zid+Did 6.91 2.00 2.9767 10.84 3.45 0.001
#>
#>
#> Inferences of model median difference between groups ( treatment 1 - treatment 0 ) for weekc = 24
#>
#> treatment 1 treatment 0 delta SE lower.CL upper.CL t.value p.value
#> 1 Zid+Zal Zid/Did 0.534 1.12 -1.660 2.73 0.479 0.632
#> 2 Zid+Did Zid/Did 4.139 1.36 1.459 6.82 3.035 0.003
#> 3 Zid+Did+Nev Zid/Did 10.135 1.66 6.874 13.40 6.108 0.000
#> 4 Zid+Did Zid+Zal 3.605 1.44 0.768 6.44 2.498 0.013
#> 5 Zid+Did+Nev Zid+Zal 9.601 1.73 6.204 13.00 5.554 0.000
#> 6 Zid+Did+Nev Zid+Did 5.996 1.89 2.291 9.70 3.180 0.002
#>
#>
#> Inferences of model median difference between groups ( treatment 1 - treatment 0 ) for weekc = 32
#>
#> treatment 1 treatment 0 delta SE lower.CL upper.CL t.value p.value
#> 1 Zid+Zal Zid/Did 1.35 1.04 -0.692 3.40 1.30 0.194
#> 2 Zid+Did Zid/Did 3.00 1.20 0.633 5.37 2.49 0.013
#> 3 Zid+Did+Nev Zid/Did 9.70 1.52 6.705 12.69 6.37 0.000
#> 4 Zid+Did Zid+Zal 1.65 1.30 -0.907 4.20 1.27 0.206
#> 5 Zid+Did+Nev Zid+Zal 8.34 1.60 5.206 11.48 5.23 0.000
#> 6 Zid+Did+Nev Zid+Did 6.70 1.71 3.338 10.06 3.92 0.000
plot(res2, ylab = "CD4+1", xlab = "Week", verbose = TRUE)
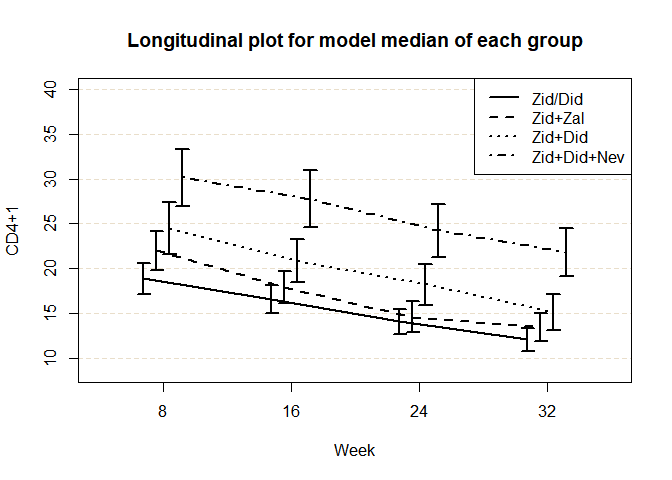
#> Analysis information:
#> Covariance structure: "UN"
#> Robust inference: TRUE
#> Empirical small sample adjustment: TRUE
#>
#> Error bar: 95% confidence interval