Tools for Modeling Bumblebee Colony Growth and Decline.
Tools for modeling bumblebee colony growth
bumbl implements a model for bumblebee colony growth described in Crone and Williams 20161. It models colony growth as having a switchpoint at some time, tau, where the colony switches from growth and worker production to gyne production and decline. Currently bumbl() works by fitting a separate switchpoint model to each colony, optimizing the switchpoint, tau. It returns the optimal switchpoint, growth, and decline rates for each colony. Because the current version of bumbl() works by fitting a separate GLM to each colony, if covariates are included, their estimates could vary significantly among colonies. Stay tuned for future developments that may allow estimating a single value of a covariate but different values of growth and decline rates and tau for each colony.
Contributing
I’m looking for collaborators who know (or are willing to let me teach them) how to use git and GitHub and who have an interest in helping to develop and maintain this package long-term. I’m not a bumblebee biologist, so I would especially love a collaborator who works on bumblebees or other organisms with a similar growth, switch, decline lifecycle.
I also welcome contributions including bug-fixes, improvement of documentation, additional features, or new functions relating to bumblebee ecology and demography from anyone!
Roadmap
- [x] Write
bumbl()function to model colony growth with switchpoint - [x] Finish documentation and vignette(s)
- [x] rOpenSci review
- [x] Regroup with Elizabeth Crone to discuss current behavior of
bumbl() - [x] Release v0.1.0 and archive on Zenodo
- [x] Bug fixes
- [x] Submit v1.0.0 of package to CRAN
- [x] Write a manuscript for JOSS (desk-reject, out of scope)
- [x] Release minor version to CRAN to update citation
- [ ] Possibly re-work internals of
bumbl()? (see #58 and notes) - [ ] Submit v2.0.0 of package to CRAN
Other possible areas of improvement:
- Extend
bumbl()to work with GLMMs - Test significance of switchpoint (see #62)
- Extend
bumbl()to work with dates, datetimes, or other timeseries classes (see #46)
Installation
You can install bumbl with:
install.packages("bumbl")
Or install the development version with:
devtools::install_github("Aariq/bumbl", build_vignettes = TRUE)
Getting started
View the package vignette with:
library(bumbl)
vignette("bumbl")
View the bomubs dataset
head(bombus)
#> # A tibble: 6 × 10
#> site colony wild habitat date week mass d.mass floral_reso…¹ cum_f…²
#> <fct> <fct> <dbl> <fct> <date> <int> <dbl> <dbl> <dbl> <dbl>
#> 1 PUT2 9 0.98 W 2003-04-03 0 1910. 0.1 27.8 27.8
#> 2 PUT2 9 0.98 W 2003-04-09 1 1940 30.6 27.8 55.5
#> 3 PUT2 9 0.98 W 2003-04-15 2 1938 28.6 27.8 83.3
#> 4 PUT2 9 0.98 W 2003-04-22 3 1976. 67.1 27.8 111.
#> 5 PUT2 9 0.98 W 2003-05-01 4 2010. 101. 7.96 119.
#> 6 PUT2 9 0.98 W 2003-05-07 5 2143 234. 7.96 127.
#> # … with abbreviated variable names ¹floral_resources, ²cum_floral
Example use
Using a subset of the bombus dataframe to estimate the week (tau) that colonies switch to reproduction
bombus2 <- bombus[bombus$colony %in% c(9, 82, 98, 35), ]
results <- bumbl(bombus2, colonyID = colony, t = week, formula = d.mass ~ week)
results
#> # A tibble: 4 × 7
#> colony converged tau logN0 logLam decay logNmax
#> <chr> <lgl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 35 TRUE 9.37 3.65 0.214 -0.296 5.60
#> 2 82 TRUE 7.34 2.91 0.407 -0.512 5.82
#> 3 9 TRUE 6.52 2.45 0.579 -0.660 6.19
#> 4 98 TRUE 6.37 1.27 0.570 -0.578 4.90
Plot the results
par(mfrow = c(2, 2))
plot(results)
#> Creating plots for 4 colonies...
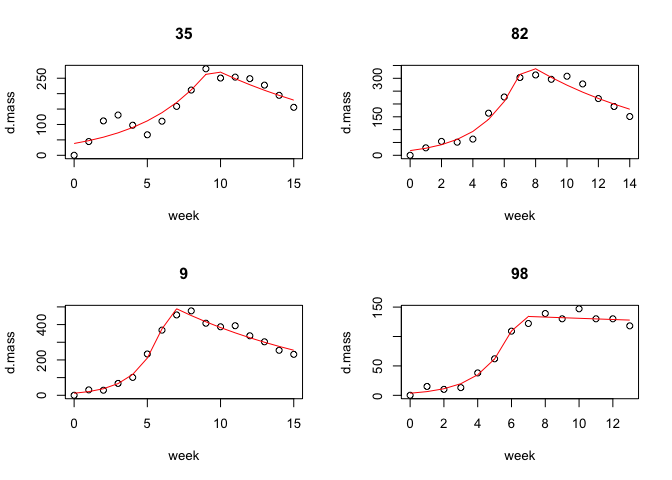
par(mfrow = c(1, 1))
References
1Crone, E. E., and Williams, N. M. (2016). Bumble bee colony dynamics: quantifying the importance of land use and floral resources for colony growth and queen production. Ecol. Lett. 19, 460–468. https://doi.org/10.1111/ele.12581
Please note that the bumbl project is released with a Contributor Code of Conduct. By contributing to this project you agree to abide by its terms.