Description
Marginal Survival Estimation with Covariate Adjustment.
Description
Provides an efficient and robust implementation for estimating marginal Hazard Ratio (HR) and Restricted Mean Survival Time (RMST) with covariate adjustment using Daniel et al. (2021) <doi:10.1002/bimj.201900297> and Karrison et al. (2018) <doi:10.1177/1740774518759281>.
README.md
bunsen: Marginal Estimation with Covariate Adjustment for Survival Endpoint in Clinical Trials
The bunsen package aims to provide an easy-to-use interface for estimating marginal or unconditional Hazard Ratio (HR) and Restricted Mean Survival Time (RMST) when adjusting prognostic covariates in clinical trials.
Key features
We introduce bunsen package for marginal HR and RMST estimation and variance for time-to-event endpoints in clinical trials. We included multiple features in current package:
- Marginal HR estimation
- Rcpp (C++) optimization
- ClusterMQ parallel computation
- Local multiprocess and multicores parallel computation for point estimate (suggested)
- LSF parallel computation for point estimate
- LSF and/or nested local multiprocess parallel computation for variance (suggested)
- RMST estimation
Package architecture
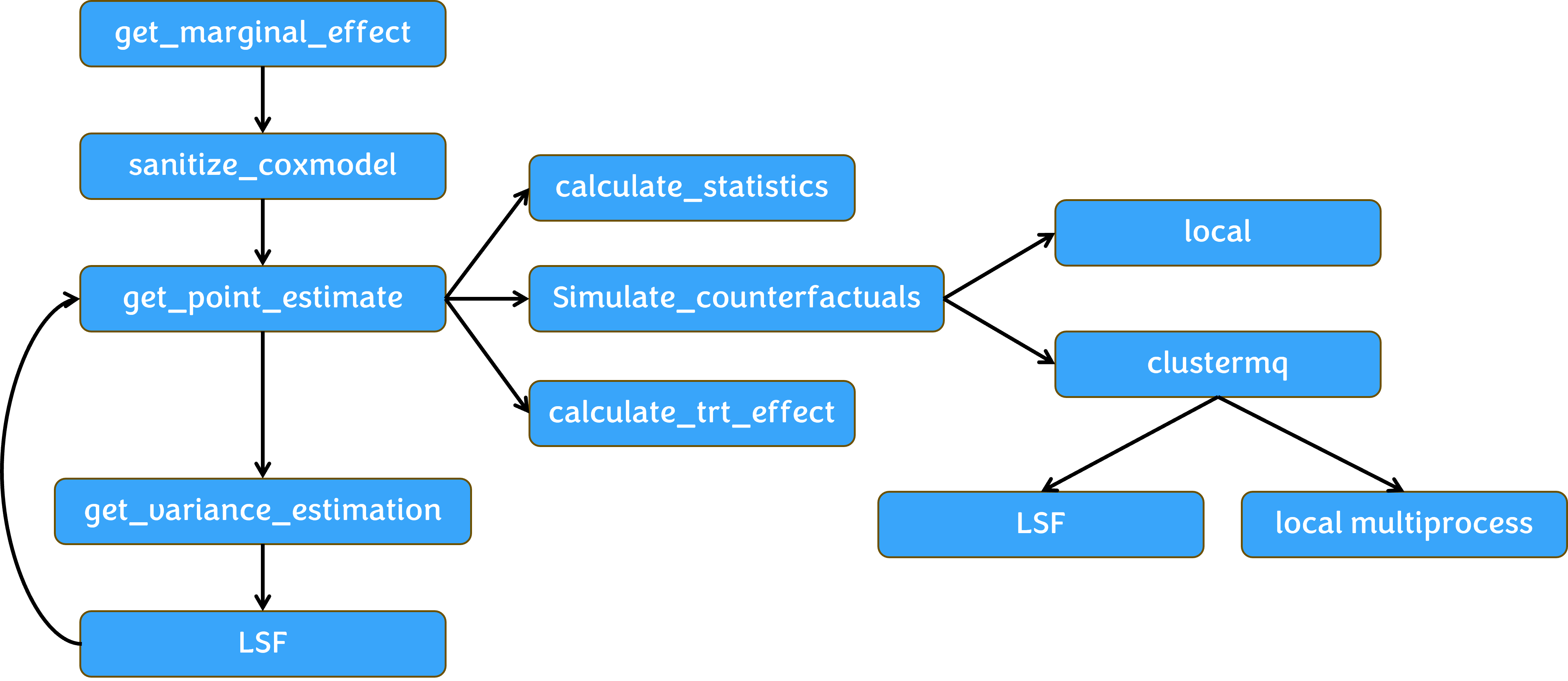
Example
Marginal point estimate and variance of HR for COX model
library(bunsen)
data('oak')
cox_event <- coxph(Surv(OS, os.status) ~ trt+btmb+pdl1, data=oak)
cox_censor <- coxph(Surv(OS, 1-os.status) ~trt+btmb+pdl1, data=oak)
result=get_marginal_effect(trt = 'trt',cox_event,cox_censor,M=10000,data=oak,seed = 1)
# Calculating point estimate in local clustermq using multiprocess...
# Submitting 4 worker jobs (ID: cmq7488) ...
# Running 4 calculations (8 objs/120.1 Kb common; 1 calls/chunk) ...
# Master: [8.6 secs 0.8% CPU]; Worker: [avg 19.1% CPU, max 303.9 Mb]
# Calculating SE in clustermq using bootstrap N = 1000...
# Submitting 100 worker jobs (ID: cmq9642) ...
# Running 1,000 calculations (14 objs/354.8 Kb common; 1 calls/chunk) ...
# Master: [16.0 secs 8.4% CPU]; Worker: [avg 48.5% CPU, max 307.7 Mb]
result
# Call:
# Surv(OS, os.status) ~ trt + btmb + pdl1
# Marginal treatment effect calculated by N = 10000 simulations
# Number of sample: 578
# coef exp(coef) se(coef) 2.5% 97.5%
# trt -0.442910 0.642165 0.103905 -0.654306 -0.245416
# clustermq setting:
# number of remote workers = 100 , each worker has 1 core(s)
# Point estimate: parallel computation (clustermq)
# SE (bootstrap): parallel computation
# 95%CI estimated by bootstrap
summary(result)
# Call:
# Surv(OS, os.status) ~ trt + btmb + pdl1
# Marginal treatment effect calculated by N = 10000 simulations
# Treatment variable: trt ------ Number of sample: 578
# Number of events in cox_event: 423
# Number of events in cox_censor: 155
# Random seed = 1
# Original treatment effect:
# coef exp(coef) se(coef) z Pr(>|z|)
# trt -0.452408 0.636095 0.098428 -4.596346 0.000004
# Marginal treatment effect:
# coef exp(coef) se(coef) z Pr(>|z|)
# trt -0.442910 0.642165 0.103905 -4.262646 0.000020
# 95% CI of Marginal treatment effect (bootstrap): -0.654 , -0.245
Marginal point estimate and variance of RMST for COX model
library(bunsen)
data('oak')
tau=26
time=oak$OS
status=oak$os.status
trt=oak$trt
covariates=oak[,c('btmb','pdl1')]
result=get_rmst_estimate(time, status, trt, covariates, tau, SE = "delta")
result
# Call:
# Surv(time, status) ~ btmb + pdl1 + strata(trt)
# Restricted survival time: 26
# coef se(coef) 2.5% 97.5%
# trt 3.265971 0.716351 1.861923 4.670019
# Method for SE calculation: delta
result=get_rmst_estimate(time, status, trt, covariates, tau, SE = "boot", seed = 2025)
result
# Call:
# Surv(time, status) ~ btmb + pdl1 + strata(trt)
# Restricted survival time: 26
# coef se(coef) 2.5% 97.5%
# trt 3.265971 0.715191 1.994362 4.699867
# Method for SE calculation: bootNumber of bootstrap: 1000 , random seed = 2025
Methodology
bunsen is developed based on three key papers:
- HR: Rhian Daniel et al. (2020)
- RMST: Theodore Karrison et al. (2018)
- Extension of above two methods: Jiawei Wei et al. (2024)
Package authors
- Xinlei Deng (Maintainer)
- Mark Baillie
- Dominic Magirr
- Craig Wang
- Alexander Przybylski
Acknowledgements
- Jiawei Wei
- Lukas Andreas Widmer.