Automatic Calibration by Evolutionary Multi Objective Algorithm.
Short Description
caRamel is an R package for optimization implementing a multiobjective evolutionary algorithm combining the MEAS algorithm and the NGSA-II algorithm.
Installation
Download and install the package from CRAN:
install.packages('caRamel')
and then load it:
library(caRamel)
Test function
Schaffer
Schaffer test function has two objectives with one variable.
Schaffer test function
schaffer <- function(i) {
if (x[i,1] <= 1) {
s1 <- -x[i,1]
} else if (x[i,1] <= 3) {
s1 <- x[i,1] - 2
} else if (x[i,1] <= 4) {
s1 <- 4 - x[i,1]
} else {
s1 <- x[i,1] - 4
}
s2 <- (x[i,1] - 5) * (x[i,1] - 5)
return(c(s1, s2))
}
Note that :
- parameter i is mandatory for the management of parallelism.
- the variable must be namedx and is a matrix of size [npopulation, nvariables].
The variable lies in the range [-5, 10]:
nvar <- 1 # number of variables
bounds <- matrix(data = 1, nrow = nvar, ncol = 2) # upper and lower bounds
bounds[, 1] <- -5 * bounds[, 1]
bounds[, 2] <- 10 * bounds[, 2]
Both functions are to be minimized:
nobj <- 2 # number of objectives
minmax <- c(FALSE, FALSE) # min and min
Before calling caRamel in order to optimize the Schaffer's problem, some algorithmic parameters need to be set:
popsize <- 100 # size of the genetic population
archsize <- 100 # size of the archive for the Pareto front
maxrun <- 1000 # maximum number of calls
prec <- matrix(1.e-3, nrow = 1, ncol = nobj) # accuracy for the convergence phase
Then the minimization problem can be launched:
results <-
caRamel(nobj,
nvar,
minmax,
bounds,
schaffer,
popsize,
archsize,
maxrun,
prec,
carallel=FALSE) # no parallelism
Test if the convergence is successful:
print(results$success==TRUE)
Plot the Pareto front:
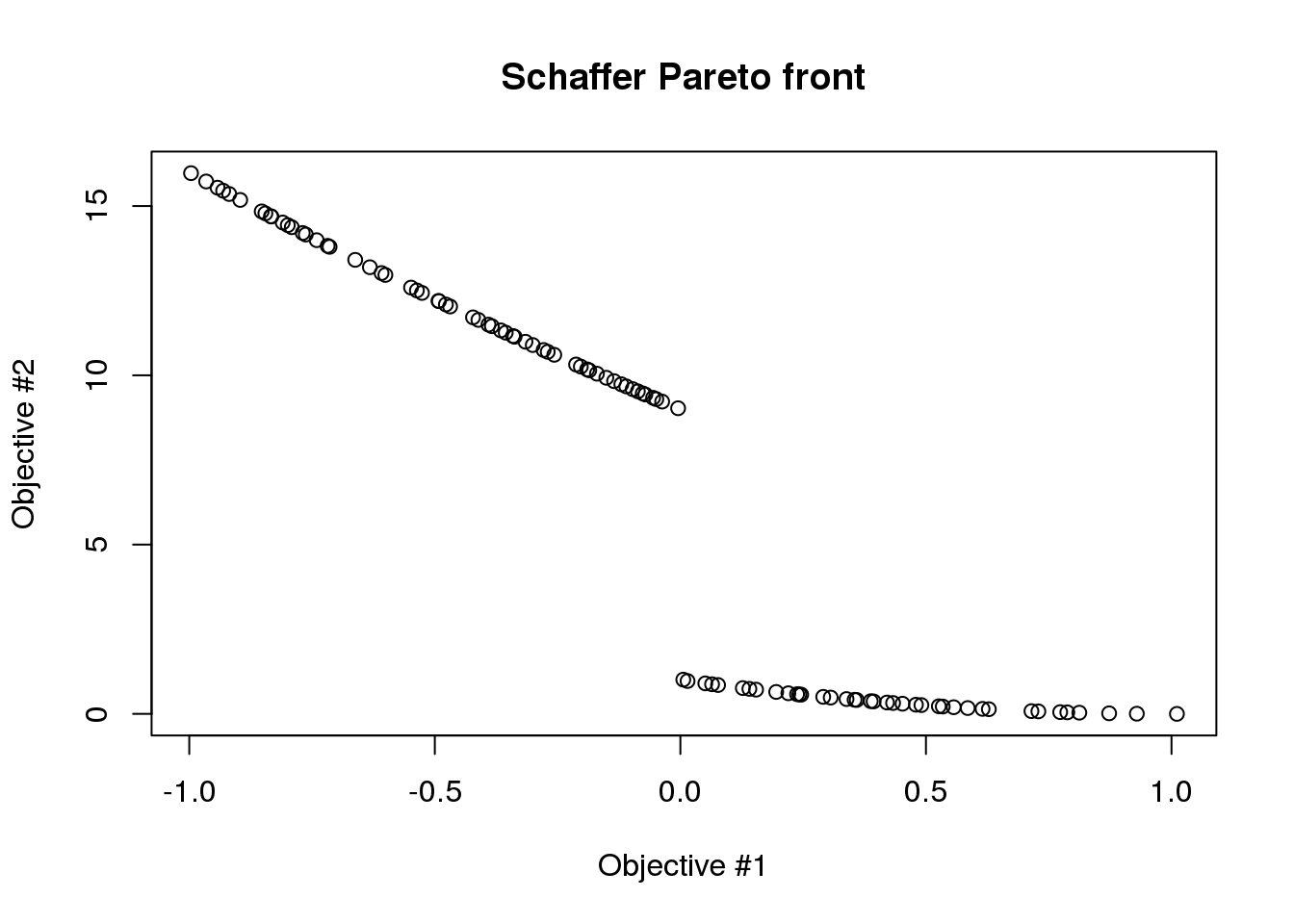
plot(results$objectives[,1], results$objectives[,2], main="Schaffer Pareto front", xlab="Objective #1", ylab="Objective #2")
plot(results$parameters, main="Corresponding values for X", xlab="Element of the archive", ylab="X Variable")
References
- Efstratiadis, A. and Koutsoyiannis, D., The multiobjective evolutionary annealing-simplex method and its application in calibrating hydrological models, EGU General Assembly 2005, Geophysical Research Abstracts, vol.7, Vienna, European Geophysical Union
- Reed, P. and Devireddy, D., Groundwater monitoring design: a case study combining epsilon-dominance archiving and automatic parameterization for the NGSA-II, Coello-Coello C editor, Applications of multiobjective evolutionary algorithms, Advances in natural computation series, vol. 1, pp. 79-100, Word Scientific, New-York, 2004
License
GPL v3
Contributors
Contributions are always welcome ;-)
When contributing to caRamel please consider discussing the changes you wish to make via issue or e-mail to the maintainer.