Description
Generic PK/PD Simulation Platform CAMPSIS.
Description
A generic, easy-to-use and intuitive pharmacokinetic/pharmacodynamic (PK/PD) simulation platform based on R packages 'rxode2' and 'mrgsolve'. CAMPSIS provides an abstraction layer over the underlying processes of writing a PK/PD model, assembling a custom dataset and running a simulation. CAMPSIS has a strong dependency to the R package 'campsismod', which allows to read/write a model from/to files and adapt it further on the fly in the R environment. Package 'campsis' allows the user to assemble a dataset in an intuitive manner. Once the user’s dataset is ready, the package is in charge of preparing the simulation, calling 'rxode2' or 'mrgsolve' (at the user's choice) and returning the results, for the given model, dataset and desired simulation settings.
README.md
campsis 
Requirements
- R package
campsismodmust be installed beforehand - Simulation engine must be installed too (
rxode2ormrgsolve)
Installation
Install the latest stable release using devtools:
devtools::install_github("Calvagone/campsis")
Basic example
Import the campsis package:
library(campsis)
Create your dataset:
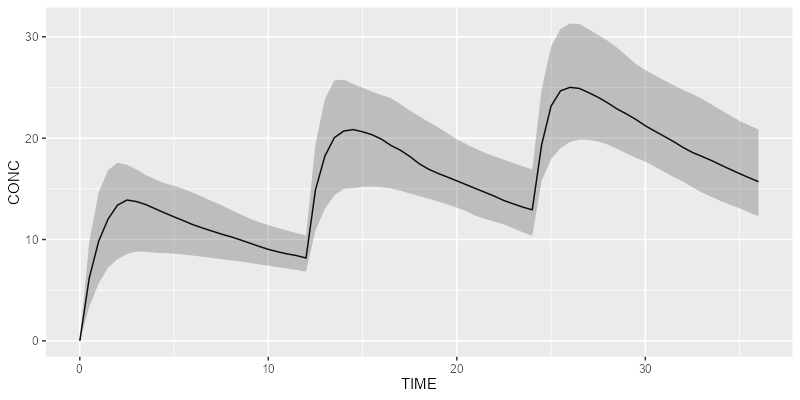
ds <- Dataset(50) %>%
add(Bolus(time=0, amount=1000, ii=12, addl=2)) %>%
add(Observations(times=seq(0, 36, by=0.5)))
Load your own model or use a built-in model from the library:
model <- model_suite$pk$`2cpt_fo`
Simulate your results with your preferred simulation engine (rxode2 or mrgsolve):
results <- model %>% simulate(dataset=ds, dest="rxode2", seed=1)
Plot your results:
shadedPlot(results, "CONC")