Description
Generic Implementation of a PK/PD Model.
Description
A generic, easy-to-use and expandable implementation of a pharmacokinetic (PK) / pharmacodynamic (PD) model based on the S4 class system. This package allows the user to read/write a pharmacometric model from/to files and adapt it further on the fly in the R environment. For this purpose, this package provides an intuitive API to add, modify or delete equations, ordinary differential equations (ODE's), model parameters or compartment properties (like infusion duration or rate, bioavailability and initial values). Finally, this package also provides a useful export of the model for use with simulation packages 'rxode2' and 'mrgsolve'. This package is designed and intended to be used with package 'campsis', a PK/PD simulation platform built on top of 'rxode2' and 'mrgsolve'.
README.md
campsismod 
Installation
Install the latest stable release using devtools:
devtools::install_github("Calvagone/campsismod")
Basic examples
Load example from model library
Load 2-compartment PK model from built-in model library:
library(campsismod)
model <- model_suite$pk$`2cpt_fo`
Write CAMPSIS model
model %>% write(file="path_to_model_folder")
list.files("path_to_model_folder")
#> [1] "model.campsis" "omega.csv" "sigma.csv" "theta.csv"
Read and show CAMPSIS model
model <- read.campsis(file="path_to_model_folder")
show(model)
#> [MAIN]
#> TVBIO=THETA_BIO
#> TVKA=THETA_KA
#> TVVC=THETA_VC
#> TVVP=THETA_VP
#> TVQ=THETA_Q
#> TVCL=THETA_CL
#>
#> BIO=TVBIO
#> KA=TVKA * exp(ETA_KA)
#> VC=TVVC * exp(ETA_VC)
#> VP=TVVP * exp(ETA_VP)
#> Q=TVQ * exp(ETA_Q)
#> CL=TVCL * exp(ETA_CL)
#>
#> [ODE]
#> d/dt(A_ABS)=-KA*A_ABS
#> d/dt(A_CENTRAL)=KA*A_ABS + Q/VP*A_PERIPHERAL - Q/VC*A_CENTRAL - CL/VC*A_CENTRAL
#> d/dt(A_PERIPHERAL)=Q/VC*A_CENTRAL - Q/VP*A_PERIPHERAL
#>
#> [F]
#> A_ABS=BIO
#>
#> [ERROR]
#> CONC=A_CENTRAL/VC
#> if (CONC <= 0.001) CONC=0.001
#> CONC_ERR=CONC*(1 + EPS_PROP_RUV)
#>
#>
#> THETA's:
#> name index value fix label unit
#> 1 BIO 1 1 FALSE Bioavailability <NA>
#> 2 KA 2 1 FALSE Absorption rate 1/h
#> 3 VC 3 10 FALSE Volume of central compartment L
#> 4 VP 4 40 FALSE Volume of peripheral compartment L
#> 5 Q 5 20 FALSE Inter-compartment flow L/h
#> 6 CL 6 3 FALSE Clearance L/h
#> OMEGA's:
#> name index index2 value fix type
#> 1 KA 1 1 25 FALSE cv%
#> 2 VC 2 2 25 FALSE cv%
#> 3 VP 3 3 25 FALSE cv%
#> 4 Q 4 4 25 FALSE cv%
#> 5 CL 5 5 25 FALSE cv%
#> SIGMA's:
#> name index index2 value fix type
#> 1 PROP_RUV 1 1 0.1 FALSE sd
#> No variance-covariance matrix
#>
#> Compartments:
#> A_ABS (CMT=1)
#> A_CENTRAL (CMT=2)
#> A_PERIPHERAL (CMT=3)
Simulate with rxode2 or mrgsolve
library(campsis)
dataset <- Dataset(5) %>%
add(Bolus(time=0, amount=1000, ii=12, addl=2)) %>%
add(Observations(times=0:36))
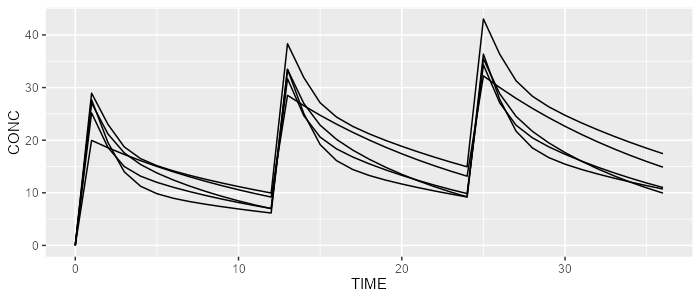
rxode <- model %>% simulate(dataset=dataset, dest="rxode2", seed=0)
mrgsolve <- model %>% simulate(dataset=dataset, dest="mrgsolve", seed=0)
spaghettiPlot(rxode, "CONC")
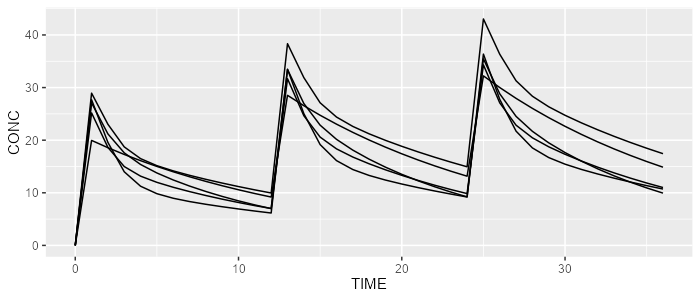
spaghettiPlot(mrgsolve, "CONC")