Carbon-Related Assessment of Silvicultural Concepts.
care4cmodel
The goal of care4cmodel is to support comparisons of different silcultural concepts on the forest estate level with a focus on the CO2 uptake by wood growth and CO2 emissions by forest operations. For achieving this, a virtual forest estate area is split into the areas covered by typical phases of the silvicultural concept of interest. Given initial area shares of these phases, the dynamics of these areas is simulated. The typical carbon stocks and flows which are known for all phases are attributed post-hoc to the areas and upscaled to the estate level. CO2 emissions by forest operations are estimated based on the amounts and dimensions of the harvested timber. Probabilities of damage events are taken into account.
Installation
In order to work with care4cmodel you require an installation of R (version >= 4.2.0) on your system. The installation files for R itself are available here. The released version of care4cmodel can be installed from CRAN with:
install.packages("care4cmodel")
Example
The following code demonstrates a basic example of how to use care4cmodel:
library(care4cmodel)
# Run a simulation and store its base results in a variable sim_base_out
# call ?simulate_single_concept for details
sim_base_out <- simulate_single_concept(
concept_def = pine_thinning_from_above_1, # use pre-defined concept
init_areas = c(800, 0, 0, 0, 0, 200),
time_span = 200,
risk_level = 3
)
# Evaluate the base results for carbon related information
# call ?fuel_and_co2_evaluation for details
carbon_out <- fuel_and_co2_evaluation(sim_base_out, road_density_m_ha = 35)
#> Joining with `by = join_by(phase_no, phase_name)`
#> Joining with `by = join_by(time, harvest_type, phase_no, phase_name)`
#> Joining with `by = join_by(time)`
# Plot results (check the documentation with ?plot.c4c_base_result, and
# ?plot.c4c_co2_result for all available options)
plot(sim_base_out) # Default plot: phase area development
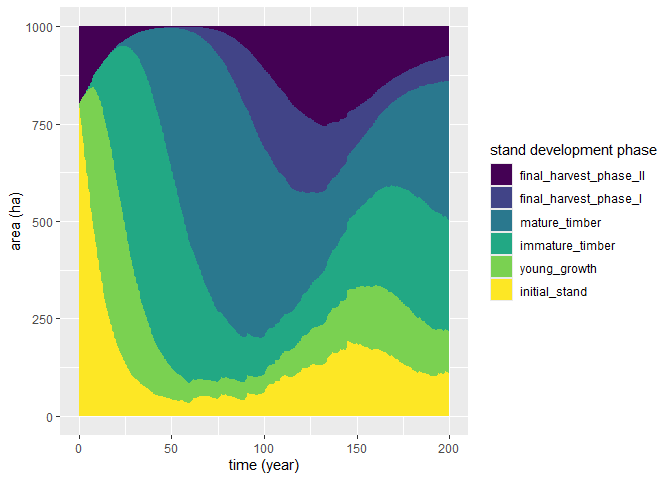
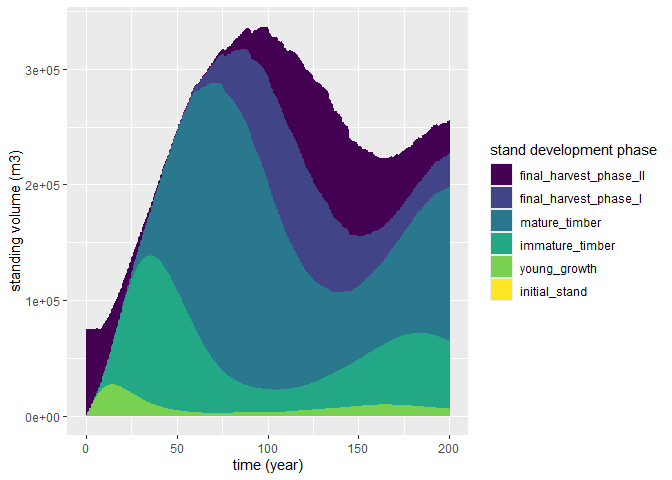
plot(sim_base_out, variable = "vol_standing") # Plot standing volume
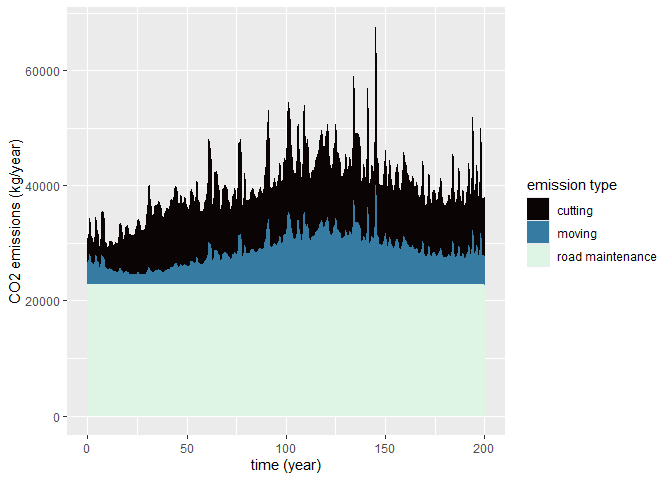
plot(carbon_out) # Default plot: CO2 emissions by emission type
More Information
For more information and details we suggest to call the package’s vignette with:
vignette("getting-started-with-care4cmodel")
A detailed scientific article about the package has been published 2024 by the authors in Computers and Electronics in Agriculture. It is available online here.
-blue.svg)