Interactively Explore Local Explanations with the Radial Tour.
cheem
Interactively explore data- and local explanation- spaces and residual side-by-side. Further explore the support of a selected observation's local explanation with the radial tour.
Context
Local explanations approximate the linear variable importance of a non-linear model in the vicinity of one instance(observation). That is, a point-measure of each variable's importance to the model at the particular location in data-space.
cheem extracts the local explanation of every observation in a dataset, given a model. Given a model, extract the local explanation of every observation in a data set. View the data- and explanation-spaces side-by-side in an interactive shiny application. Further explored a selected point against a comparison using its explanation as a 1D projection basis. A radial tour then explores the structure of explanation projection.
Getting started
## Download the package
install.packages("cheem", dependencies = TRUE)
## May need to restart the R session so RSudio has the correct file structure
rstudioapi::restartSession()
## Load cheem into session
library(cheem)
## Try the app
run_app()
## Processing your data; follow the examples in cheem_ls()
?cheem_ls
Global view
The global view shows data-, attribution-spaces, and residual plot side-by-side with linked brushing and hover tooltip.
By exploring the global view, identify a primary and comparison observation to compare. For the classification task, typically a misclassified point is selected and compared against a nearby correctly classified one. In regression, we can compare a point with an extreme residual with a nearby point that is more accurately predicted.
Radial cheem tour
The attribution of the primary observation becomes the 1D basis for the tour. The variable with the largest difference between the primary and comparison point's bases is selected as the manipulation variable. That is the variable whose contribution change drives the change in the projection basis.
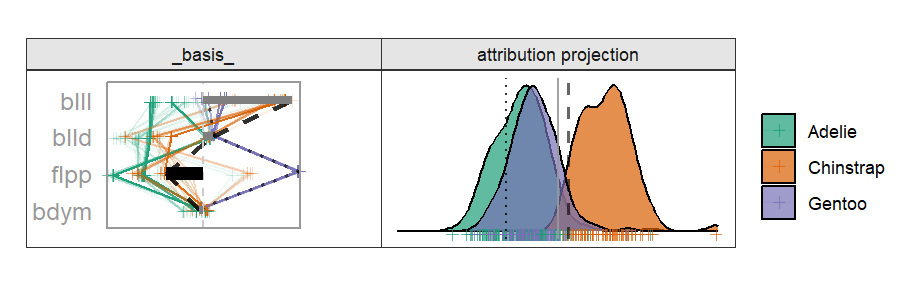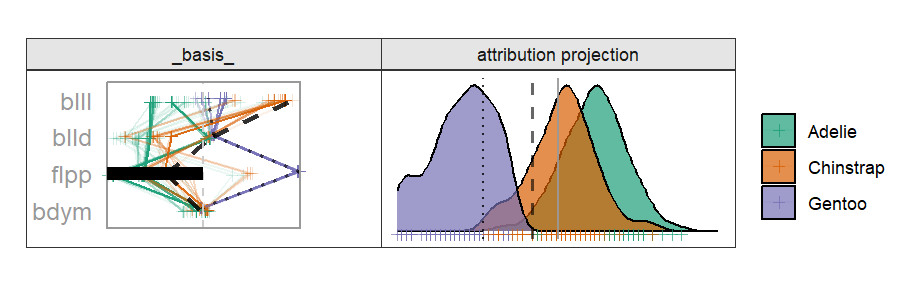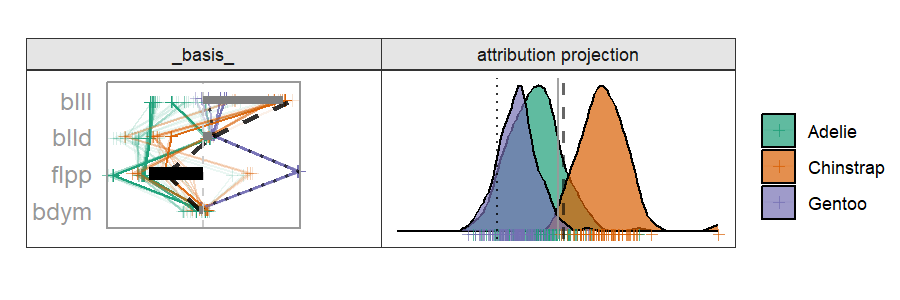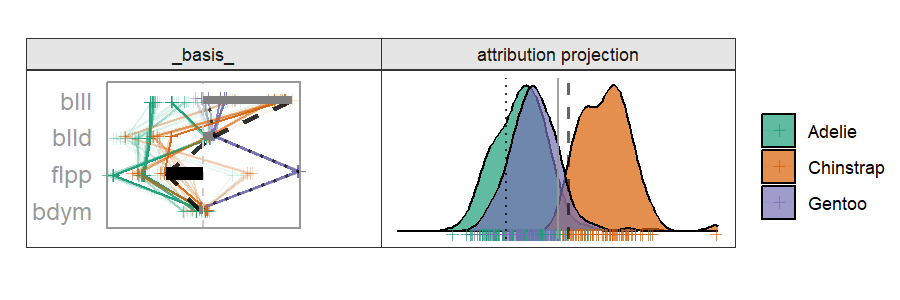
By doing this, we are testing the local explanation. By testing the variable sensitivity to the structure identified in the local explanation, we can better evaluate how good of an explanation it is; how sensitive its prediction is to a change in the variable contributions.
Original application
We started by looking at the model-agnostic local explanation tree SHAP applied to random forests. We made this choice out of concern for runtime (treeshap uses an alternative algorithm with reduced computational complexity and thus achieves much faster run time extracting the full SHAP matrix during the preprocessing step). The namesake, Cheem, stems from the original application to tree-based models in the DALEX ecosystem; Cheem are a fictional race of tree-based humanoids for consistency with the Dr. who/Dr. why theme.
Sources
Package build workflow
- devtools::document() ## documentation changes
- pkgdown::build_site() ## packagedown site changes (documentation, vignettes, readme)
- message("Manually do: Build tab > Install and Restart") ## build package
- rhub::check_for_cran() ## check package
- devtools::submit_cran() ## Submit to CRAN.