Chemical Metrics for Microbial Communities.
chem16S generates chemical representations of microbial communities by combining taxonomic abundances of archaea and bacteria with reference sequences for proteins. Chemical metrics for community reference proteomes are used to investigate genomic adaptations to environmental conditions. Potential applications range from human microbiomes to Earth-life coevolution.
Read the paper in Bioinformatics: chem16S: community-level chemical metrics for exploring genomic adaptation to environments.
View the manual and vignettes: Chemical metrics of reference proteomes for taxa, Integration of chem16S with phyloseq, and Plotting two chemical metrics.
Methods
The user provides taxonomic classifications of high-throughput 16S rRNA gene sequences. These are combined with reference proteomes for archaea and bacteria to obtain the amino acid compositions of community reference proteomes. Amino acid compositions used to calculate chemical metrics including carbon oxidation state (Z/sub) and stoichiometric hydration state (nH2O).
Supported input formats:
phyloseq-classobjects created using phyloseq- RDP Classifier output
Supported reference databases:
- Genome Taxonomy Database (GTDB release 220)
- NCBI Reference Sequence Database (RefSeq release 206)
- Scripts used to generate reference proteomes for genus- and higher-level archaeal and bacterial taxa (and viruses for RefSeq) are available in the GTDB_220 and RefSeq_206 directories.
Details:
- The taxonomic classifier should be trained on 16S rRNA sequences from GTDB so that taxon names are matched to GTDB reference proteomes available in chem16S. Training files are available for DADA2 and the RDP Classifier.
- For taxonomic classifications made using the RDP training set (No. 18 07/2020, used in RDP Classifier version 2.13), chem16S includes manual mappings to the NCBI taxonomy described by Dick and Tan (2023).
Example
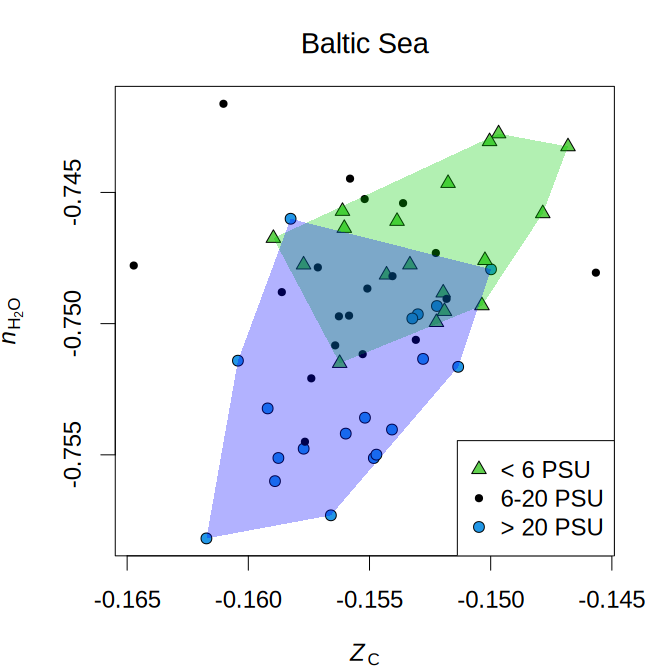
The Baltic Sea has a salinity gradient from freshwater to marine conditions. Progressively lower nH2O of community reference proteomes along this gradient represent a genomically coded dehydration trend.
PSU stands for practical salinity units. The sequence data analyzed for this plot was taken from Herlemann et al. (2016) and the code to make this plot is available in the help page for chem16S::plot_metrics.
Installation
First install phyloseq from Bioconductor:
if(!require("BiocManager", quietly = TRUE)) install.packages("BiocManager")
BiocManager::install("phyloseq")
Then install the release version of chem16S from CRAN:
install.packages("chem16S")
Or use install_github from remotes or devtools to install the development version of chem16S from GitHub:
if(!require("remotes", quietly = TRUE)) install.packages("remotes")
remotes::install_github("jedick/chem16S", build_vignettes = TRUE)