Description
A Computational Pipeline for Bulk 'ATAC-Seq' Profiles.
Description
Differential analyses and Enrichment pipeline for bulk 'ATAC-seq' data analyses. This package combines different packages to have an ultimate package for both data analyses and visualization of 'ATAC-seq' data. Methods are described in 'Karakaslar et al.' (2021) <doi:10.1101/2021.03.05.434143>.
README.md
cinaR 
Overview
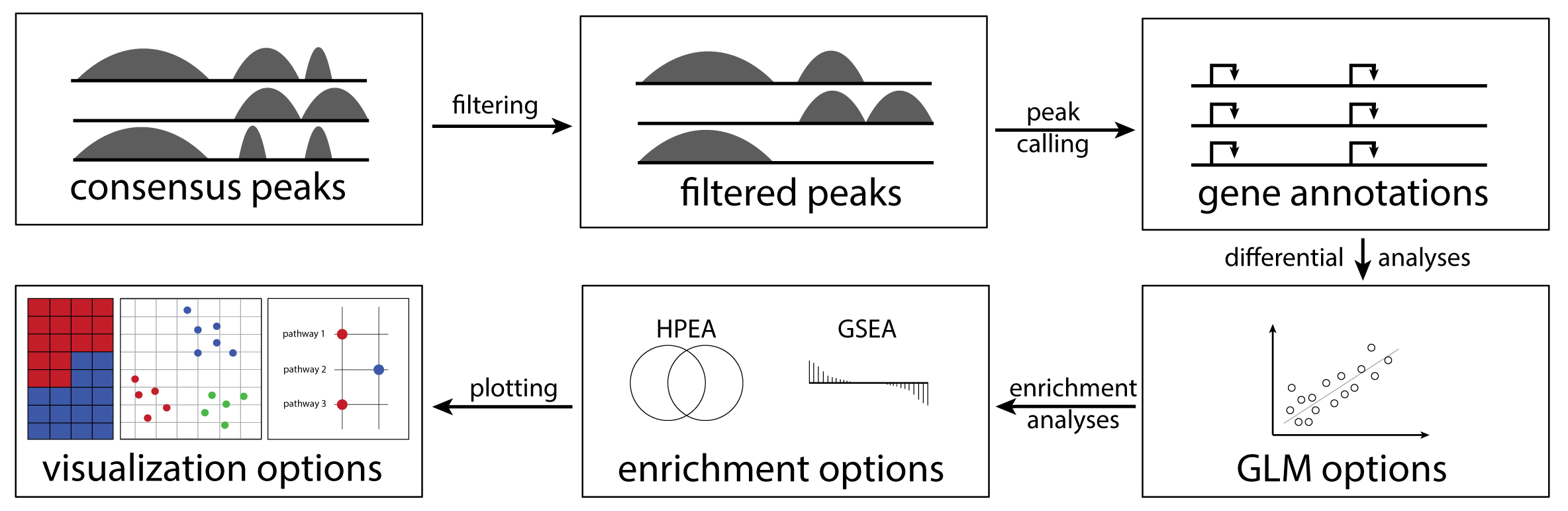
cinaR is a single wrapper function for end-to-end computational analyses of bulk ATAC-seq (or RNA-seq) profiles. Starting from a consensus peak file, it outputs differentially accessible peaks, enrichment results, and provides users with various configurable visualization options. For more details, please see the preprint.
Installation
# CRAN mirror
install.packages("cinaR")
Development version
To get bug fix and use a feature from the development version:
# install.packages("devtools")
devtools::install_github("eonurk/cinaR")
Usage
For more details please go to our site from here!
Known Installation Issues
Sometimes bioconductor related packages may not be installed automatically.
Therefore, you may need to install them manually:
BiocManager::install(c("ChIPseeker", "DESeq2", "edgeR", "fgsea","GenomicRanges", "limma", "preprocessCore", "sva", "TxDb.Hsapiens.UCSC.hg38.knownGene", "TxDb.Hsapiens.UCSC.hg19.knownGene", "TxDb.Mmusculus.UCSC.mm10.knownGene"))
Contribution
You can send pull requests to make your contributions.
Author
License
- GNU General Public License v3.0