Simplifies Access to Cloudstor API.
cloudstoR
The goal of cloudstoR is to simplify accessing data stored on Cloudstor via their WebDAV interface. You can use cloudstoR to download or upload files, or check the contents of directories.
Installation
You can install from CRAN with:
install.packages("cloudstoR")
Or install the development version from GitHub with:
# install.packages("devtools")
devtools::install_github("pdparker/cloudstoR")
For additional support for the package, check the pkgdown site.
Setup
You will need your Cloudstor username and password. The password is not the one you use to log on to Cloudstor. Instead you need to use an app password. cloudstoR provides an option to store these credentials using keyring.
Examples
Setting credentials
The first time you use a cloud_* function, cloudstoR will prompt you to store your credentials locally. You can choose not to do this by providing a username and password for each function call.
library(cloudstoR)
my_data <- cloud_get(path = 'cloudstoR Tests/mydata1.csv',
username = cloudstor_username,
password = cloudstor_appPassword)
Getting a list of files
To retrieve a list of files in a folder, use cloud_list(). The path argument specifies the folder to return.
cloud_list(path = 'cloudstoR Tests')
#> [1] "Another Folder/" "mydata1.csv" "mydata2.csv"
Getting a specific file
To retrieve a file use cloud_get(), and provide the path to the file with the path argument.
my_data <- cloud_get(path = 'cloudstoR Tests/mydata1.csv')
my_data
#> A B C
#> 1 3 10 8
#> 2 5 7 5
#> 3 5 7 10
By default, cloudstoR will try to open the file using rio and return a data.frame. The temporary version of the file is deleted once it is read into memory. If you want to use a different package to open the file, or you just want to download and keep the file without opening it, set open_file = FALSE to return a file path instead.
my_path <- cloud_get(path = 'cloudstoR Tests/mydata1.csv',
dest = "~/mydata1.csv",
open_file = FALSE)
file.exists(my_path)
#> [1] TRUE
Saving a file to Cloudstor
To upload a file, use cloud_put(). You need to provide the path to the saved file (local_file), and the path to save the file on Cloudstor (path). You can optionally provide a name for the file (file_name), otherwise the file name of the local file is used.
cloud_put(local_file = '~/datatosave.sav',
path = 'additional/path/to/folder',
file_name = 'mydata.sav',)
Navigating the folder tree
If you don’t know the exact file path you want to follow, you can find it with cloud_browse().
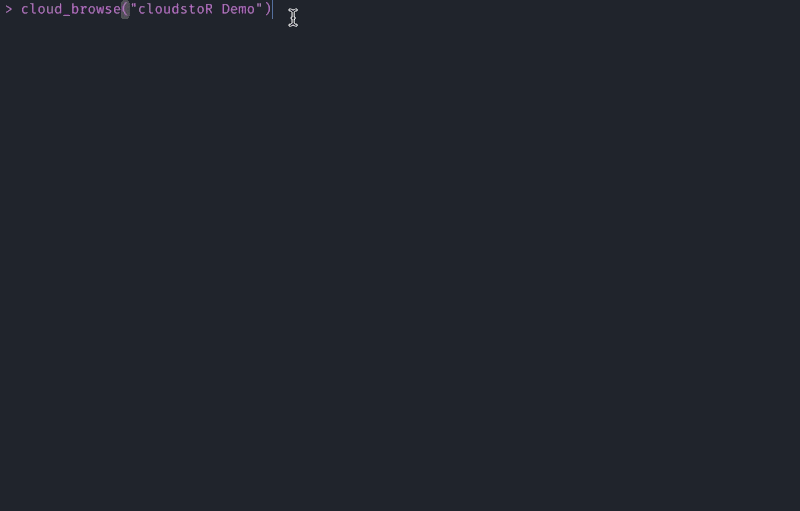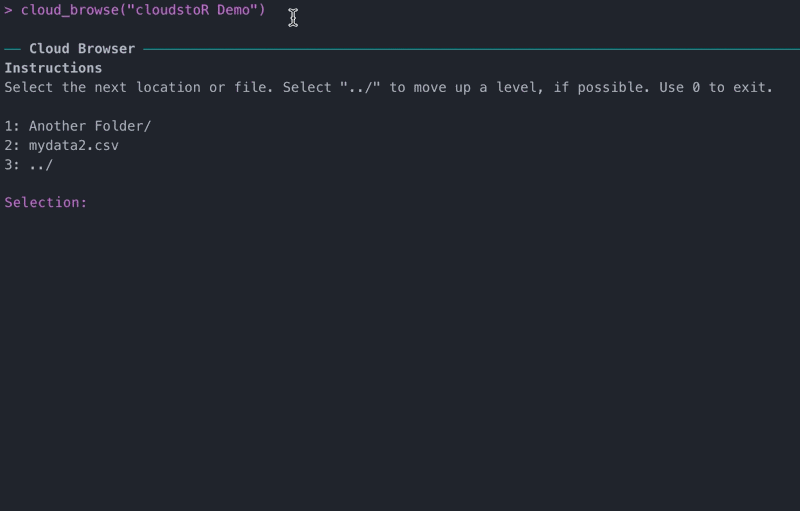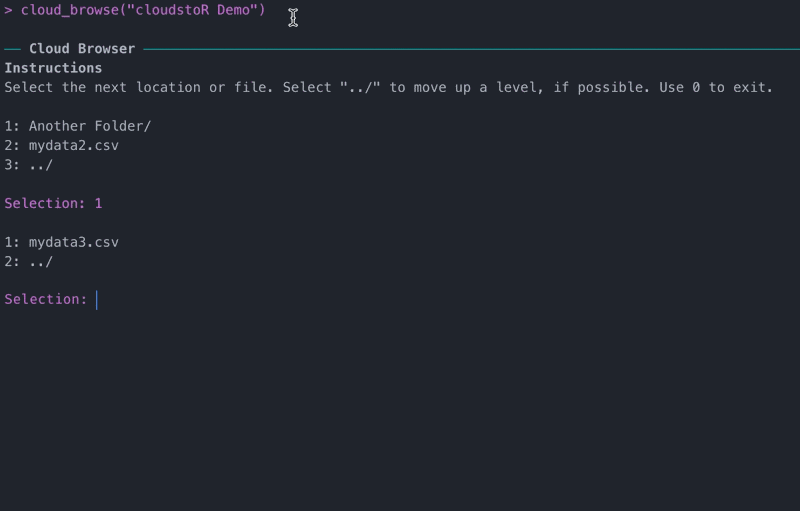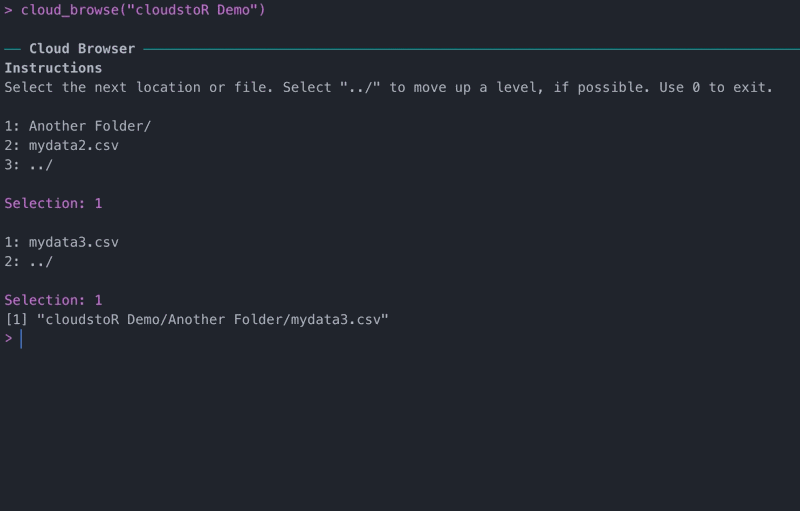
View file or folder meta-data
You can view meta-data for a file or folder with cloud_meta(). This can be especially useful for checking if a file has been modified.
cloud_meta(path = 'cloudstoR Tests/mydata1.csv')
#> file_name
#> 1 /plus/remote.php/webdav/cloudstoR Tests/mydata1.csv
#> tag file_modified file_size
#> 1 "9a2a8fdd58a6c2746cd65b7dace6115c" Sun, 16 Jan 2022 05:42:52 GMT 36
Updating credentials
If you need to delete your credentials (e.g., because you revoke your app password), you can restore them by calling cloud_auth() directly:
cloud_auth(reset_keys=TRUE)
Using an alternative WebDAV address
The default WebDAV address is https://cloudstor.aarnet.edu.au/plus/remote.php/webdav/. If your organisation uses a different address, you can set this globally at the top of your script:
# Set the global WebDAV address
options(cloudstoR.cloud_address = "https:://my.webdav.address")
# Check the current value
getOption("cloudstoR.cloud_address")
