Cluster Strings by Edit-Distance.
clustringr
clustringr clusters a vector of strings into groups of small mutual "edit distance" (see stringdist), using graph algorithms. Notice it's unsupervised, i.e., you do not need to pre-specify cluster count. Graph visualization of the results is provided.
Installation
Currently a development version is available on github.
# install.packages('devtools')
devtools::install_github('dan-reznik/clustringr')
Usage
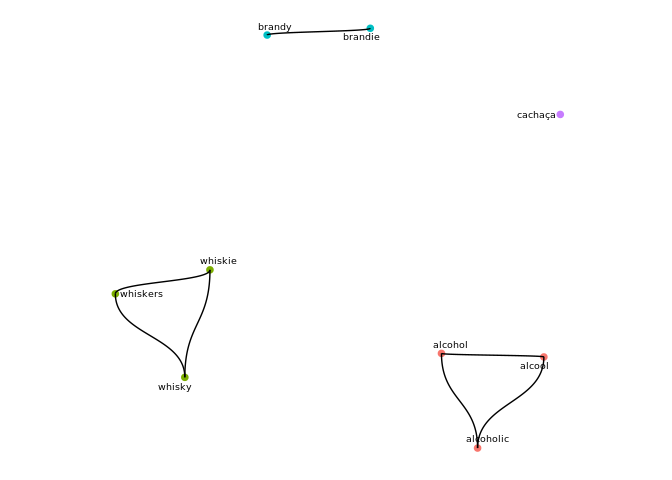
In the example below a vector of 9 strings is clustered into 4 groups by levenshtein distance and connected components. The call to cluster_strings() returns a list w/ 3 elements, the last of which is df_clusters which associates to every input string a cluster, along with its cluster size.
library(clustringr)
s_vec <- c("alcool",
"alcohol",
"alcoholic",
"brandy",
"brandie",
"cachaça",
"whisky",
"whiskie",
"whiskers")
s_clust <- cluster_strings(s_vec # input vector
,clean=T # dedup and squish
,method="lv" # levenshtein
# use: method="dl" (dam-lev) or "osa" for opt-seq-align
,max_dist=3 # max edit distance for neighbors
,algo="cc" # connected components
# use algo="eb" for edge-betweeness
)
s_clust$df_clusters
#> # A tibble: 9 x 3
#> cluster size node
#> <int> <int> <chr>
#> 1 1 3 alcohol
#> 2 1 3 alcoholic
#> 3 1 3 alcool
#> 4 2 3 whiskers
#> 5 2 3 whiskie
#> 6 2 3 whisky
#> 7 3 2 brandie
#> 8 3 2 brandy
#> 9 4 1 cachaça
Cluster Visualization
To view a graph of the clusters, simply pass the structure returned by cluster_strings to cluster_plot:
cluster_plot(s_clust
,min_cluster_size=1
# ,label_size=2.5 # size of node labels
# ,repel=T # whether labels should be repelled
)
#> Using `nicely` as default layout
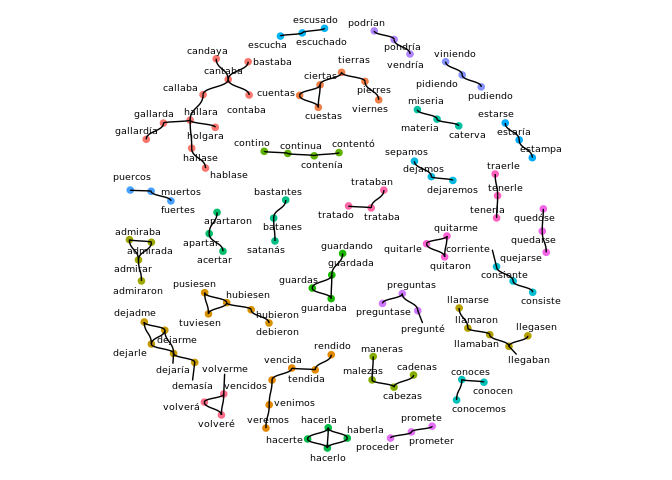
Supplied Data Set: Don Quijote's unique words
The clustringr package comes with quijote_words, a ~22k row data frame of the unique words (in Spanish) in Miguel de Cervantes' "Don Quijote". Full text can be obtained here.
Let's sample these words into a smaller subset:
library(dplyr)
quijote_words_sampled <- clustringr::quijote_words %>%
filter(between(freq,8,11),len>6) %>%
pull("word")
quijote_words_sampled%>%length
#> [1] 602
Now let's cluster these and view the results as a graph-plot, showing only those clusters with at least 3 elements:
quijote_words_sampled %>%
cluster_strings(method="lv",max_dist=2) %>%
cluster_plot(min_cluster_size=3)
Happy clustering!