Data Source Agnostic Filtering Tools.
cohortBuilder 
Overview
cohortBuilder provides common API for creating cohorts on multiple data sources, such as local data frame, database schema or external data api.
With only two steps:
- Configuring data source with
set_source. - Initializing cohort with
cohort.
You can operate on data using common methods, such as:
filter- to define andrunto apply filtering rules,step- to perform multi-stage filtering,get_data,stat,attrition,plot_data- to extract, sum up or visualize your cohort data.
With cohortBuilder you can share the cohort easier with useful methods:
code- to get reproducible cohort creation code,get_state- to get cohort state (e.g. in JSON) that can be then easily restored withrestore.
Or modify the cohort configuration with:
add_filter,rm_filter,update_filter- to manage filters definitionadd_step,rm_step- to manage filtering steps,update_source- to manage the cohort source.
Data sources and extensions
The goal of cohortBuilder is to provide common API for creating data cohorts, but also to be easily extendable for working on different data sources (and interactive dashboards).
cohortBuilder allows to operate on local data frames (or list of data frames), yet you may easily switch to a database source by loading cohortBuilder.db layer.
As a standalone R package, you use cohortBuilder to perform all the operations in non-interactive R script, but its shiny layer shinyCohortBuilder package helps you to easily switch to intuitive gui mode. More to that you may integrate cohortBuilder with your custom Shiny application.
If you want to learn how to write custom source extension, please check vignette("custom-extensions").
Installation
# CRAN version
install.packages("cohortBuilder")
# Latest development version
remotes::install_github("https://github.com/r-world-devs/cohortBuilder")
Usage
librarian_source <- set_source(
as.tblist(librarian)
)
coh <- librarian_source %>%
cohort(
filter(
"discrete", id = "author", dataset = "books",
variable = "author", value = "Dan Brown"
),
filter(
"range", id = "copies", dataset = "books",
variable = "copies", range = c(5, 10)
),
filter(
"date_range", id = "registered", dataset = "borrowers",
variable = "registered", range = c(as.Date("2010-01-01"), Inf)
)
) %>%
run()
get_data(coh)
#> $books
#> # A tibble: 1 × 6
#> isbn title genre publisher
#> <chr> <chr> <chr> <chr>
#> 1 0-385-50420-9 The Da Vinci Code Crime, Thriller & Adventure Transworld
#> author copies
#> <chr> <int>
#> 1 Dan Brown 7
#>
#> $borrowers
#> # A tibble: 8 × 6
#> id registered address
#> <chr> <date> <chr>
#> 1 000013 2011-09-30 534 Iroquois Ave. Watertown, MA 02472
#> 2 000014 2013-01-12 7968 Victoria Drive Dearborn, MI 48124
#> 3 000015 2013-12-24 9484 Somerset Road Romeoville, IL 60446
#> 4 000016 2014-01-20 48 Prairie Ave. Palos Verdes Peninsula, CA 90274
#> 5 000017 2014-04-07 8501 Lawrence Rd. Terre Haute, IN 47802
#> name phone_number program
#> <chr> <chr> <chr>
#> 1 Dr. Sharif Kunde 104-832-8013 premium
#> 2 Marlena Reichert PhD 044-876-8419 vip
#> 3 Mr. Brandan Oberbrunner 568-044-7463 vip
#> 4 Lloyd Adams III 001-017-0211 standard
#> 5 Randy Ziemann 895-995-2326 premium
#> # ℹ 3 more rows
#>
#> $issues
#> # A tibble: 50 × 4
#> id borrower_id isbn date
#> <chr> <chr> <chr> <date>
#> 1 000001 000019 0-676-97976-9 2015-03-17
#> 2 000002 000010 978-0-7528-6053-4 2008-09-13
#> 3 000003 000016 0-09-177373-3 2014-09-28
#> 4 000004 000005 0-224-06252-2 2005-11-14
#> 5 000005 000004 0-340-89696-5 2006-03-19
#> # ℹ 45 more rows
#>
#> $returns
#> # A tibble: 30 × 2
#> id date
#> <chr> <date>
#> 1 000001 2015-04-06
#> 2 000003 2014-10-23
#> 3 000004 2005-12-29
#> 4 000005 2006-03-26
#> 5 000006 2016-08-30
#> # ℹ 25 more rows
#>
#> attr(,"class")
#> [1] "tblist"
#> attr(,"call")
#> as.tblist(librarian)
coh <- librarian_source %>%
cohort() %->%
step(
filter(
"discrete", id = "author", dataset = "books",
variable = "author", value = "Dan Brown"
),
filter(
"date_range", id = "registered", dataset = "borrowers",
variable = "registered", range = c(as.Date("2010-01-01"), Inf)
)
) %->%
step(
filter(
"range", id = "copies", dataset = "books",
variable = "copies", range = c(5, 10)
)
) %>%
run()
get_data(coh, step_id = 1)
#> $books
#> # A tibble: 2 × 6
#> isbn title genre publisher
#> <chr> <chr> <chr> <chr>
#> 1 0-385-50420-9 The Da Vinci Code Crime, Thriller & Adventure Transworld
#> 2 0-671-02735-2 Angels and Demons Crime, Thriller & Adventure Transworld
#> author copies
#> <chr> <int>
#> 1 Dan Brown 7
#> 2 Dan Brown 4
#>
#> $borrowers
#> # A tibble: 8 × 6
#> id registered address
#> <chr> <date> <chr>
#> 1 000013 2011-09-30 534 Iroquois Ave. Watertown, MA 02472
#> 2 000014 2013-01-12 7968 Victoria Drive Dearborn, MI 48124
#> 3 000015 2013-12-24 9484 Somerset Road Romeoville, IL 60446
#> 4 000016 2014-01-20 48 Prairie Ave. Palos Verdes Peninsula, CA 90274
#> 5 000017 2014-04-07 8501 Lawrence Rd. Terre Haute, IN 47802
#> name phone_number program
#> <chr> <chr> <chr>
#> 1 Dr. Sharif Kunde 104-832-8013 premium
#> 2 Marlena Reichert PhD 044-876-8419 vip
#> 3 Mr. Brandan Oberbrunner 568-044-7463 vip
#> 4 Lloyd Adams III 001-017-0211 standard
#> 5 Randy Ziemann 895-995-2326 premium
#> # ℹ 3 more rows
#>
#> $issues
#> # A tibble: 50 × 4
#> id borrower_id isbn date
#> <chr> <chr> <chr> <date>
#> 1 000001 000019 0-676-97976-9 2015-03-17
#> 2 000002 000010 978-0-7528-6053-4 2008-09-13
#> 3 000003 000016 0-09-177373-3 2014-09-28
#> 4 000004 000005 0-224-06252-2 2005-11-14
#> 5 000005 000004 0-340-89696-5 2006-03-19
#> # ℹ 45 more rows
#>
#> $returns
#> # A tibble: 30 × 2
#> id date
#> <chr> <date>
#> 1 000001 2015-04-06
#> 2 000003 2014-10-23
#> 3 000004 2005-12-29
#> 4 000005 2006-03-26
#> 5 000006 2016-08-30
#> # ℹ 25 more rows
#>
#> attr(,"class")
#> [1] "tblist"
#> attr(,"call")
#> as.tblist(librarian)
get_data(coh, step_id = 2)
#> $books
#> # A tibble: 1 × 6
#> isbn title genre publisher
#> <chr> <chr> <chr> <chr>
#> 1 0-385-50420-9 The Da Vinci Code Crime, Thriller & Adventure Transworld
#> author copies
#> <chr> <int>
#> 1 Dan Brown 7
#>
#> $borrowers
#> # A tibble: 8 × 6
#> id registered address
#> <chr> <date> <chr>
#> 1 000013 2011-09-30 534 Iroquois Ave. Watertown, MA 02472
#> 2 000014 2013-01-12 7968 Victoria Drive Dearborn, MI 48124
#> 3 000015 2013-12-24 9484 Somerset Road Romeoville, IL 60446
#> 4 000016 2014-01-20 48 Prairie Ave. Palos Verdes Peninsula, CA 90274
#> 5 000017 2014-04-07 8501 Lawrence Rd. Terre Haute, IN 47802
#> name phone_number program
#> <chr> <chr> <chr>
#> 1 Dr. Sharif Kunde 104-832-8013 premium
#> 2 Marlena Reichert PhD 044-876-8419 vip
#> 3 Mr. Brandan Oberbrunner 568-044-7463 vip
#> 4 Lloyd Adams III 001-017-0211 standard
#> 5 Randy Ziemann 895-995-2326 premium
#> # ℹ 3 more rows
#>
#> $issues
#> # A tibble: 50 × 4
#> id borrower_id isbn date
#> <chr> <chr> <chr> <date>
#> 1 000001 000019 0-676-97976-9 2015-03-17
#> 2 000002 000010 978-0-7528-6053-4 2008-09-13
#> 3 000003 000016 0-09-177373-3 2014-09-28
#> 4 000004 000005 0-224-06252-2 2005-11-14
#> 5 000005 000004 0-340-89696-5 2006-03-19
#> # ℹ 45 more rows
#>
#> $returns
#> # A tibble: 30 × 2
#> id date
#> <chr> <date>
#> 1 000001 2015-04-06
#> 2 000003 2014-10-23
#> 3 000004 2005-12-29
#> 4 000005 2006-03-26
#> 5 000006 2016-08-30
#> # ℹ 25 more rows
#>
#> attr(,"class")
#> [1] "tblist"
#> attr(,"call")
#> as.tblist(librarian)
update_filter(
coh, step_id = 1, filter_id = "author",
range = c(5, 6)
)
run(coh)
get_data(coh, step_id = 2)
#> $books
#> # A tibble: 1 × 6
#> isbn title genre publisher
#> <chr> <chr> <chr> <chr>
#> 1 0-385-50420-9 The Da Vinci Code Crime, Thriller & Adventure Transworld
#> author copies
#> <chr> <int>
#> 1 Dan Brown 7
#>
#> $borrowers
#> # A tibble: 8 × 6
#> id registered address
#> <chr> <date> <chr>
#> 1 000013 2011-09-30 534 Iroquois Ave. Watertown, MA 02472
#> 2 000014 2013-01-12 7968 Victoria Drive Dearborn, MI 48124
#> 3 000015 2013-12-24 9484 Somerset Road Romeoville, IL 60446
#> 4 000016 2014-01-20 48 Prairie Ave. Palos Verdes Peninsula, CA 90274
#> 5 000017 2014-04-07 8501 Lawrence Rd. Terre Haute, IN 47802
#> name phone_number program
#> <chr> <chr> <chr>
#> 1 Dr. Sharif Kunde 104-832-8013 premium
#> 2 Marlena Reichert PhD 044-876-8419 vip
#> 3 Mr. Brandan Oberbrunner 568-044-7463 vip
#> 4 Lloyd Adams III 001-017-0211 standard
#> 5 Randy Ziemann 895-995-2326 premium
#> # ℹ 3 more rows
#>
#> $issues
#> # A tibble: 50 × 4
#> id borrower_id isbn date
#> <chr> <chr> <chr> <date>
#> 1 000001 000019 0-676-97976-9 2015-03-17
#> 2 000002 000010 978-0-7528-6053-4 2008-09-13
#> 3 000003 000016 0-09-177373-3 2014-09-28
#> 4 000004 000005 0-224-06252-2 2005-11-14
#> 5 000005 000004 0-340-89696-5 2006-03-19
#> # ℹ 45 more rows
#>
#> $returns
#> # A tibble: 30 × 2
#> id date
#> <chr> <date>
#> 1 000001 2015-04-06
#> 2 000003 2014-10-23
#> 3 000004 2005-12-29
#> 4 000005 2006-03-26
#> 5 000006 2016-08-30
#> # ℹ 25 more rows
#>
#> attr(,"class")
#> [1] "tblist"
#> attr(,"call")
#> as.tblist(librarian)
code(coh)
#> .pre_filtering <- function(source, data_object, step_id) {
#> for (dataset in names(data_object)) {
#> attr(data_object[[dataset]], "filtered") <- FALSE
#> }
#> return(data_object)
#> }
#> .run_binding <- function(source, binding_key, data_object_pre, data_object_post,
#> ...) {
#> binding_dataset <- binding_key$update$dataset
#> dependent_datasets <- names(binding_key$data_keys)
#> active_datasets <- data_object_post %>%
#> purrr::keep(~attr(., "filtered")) %>%
#> names()
#> if (!any(dependent_datasets %in% active_datasets)) {
#> return(data_object_post)
#> }
#> key_values <- NULL
#> common_key_names <- paste0("key_", seq_along(binding_key$data_keys[[1]]$key))
#> for (dependent_dataset in dependent_datasets) {
#> key_names <- binding_key$data_keys[[dependent_dataset]]$key
#> tmp_key_values <- dplyr::distinct(data_object_post[[dependent_dataset]][,
#> key_names, drop = FALSE]) %>%
#> stats::setNames(common_key_names)
#> if (is.null(key_values)) {
#> key_values <- tmp_key_values
#> } else {
#> key_values <- dplyr::inner_join(key_values, tmp_key_values, by = common_key_names)
#> }
#> }
#> data_object_post[[binding_dataset]] <- dplyr::inner_join(switch(as.character(binding_key$post),
#> `FALSE` = data_object_pre[[binding_dataset]], `TRUE` = data_object_post[[binding_dataset]]),
#> key_values, by = stats::setNames(common_key_names, binding_key$update$key))
#> if (binding_key$activate) {
#> attr(data_object_post[[binding_dataset]], "filtered") <- TRUE
#> }
#> return(data_object_post)
#> }
#> source <- list(dtconn = as.tblist(librarian))
#> data_object <- source$dtconn
#> step_id <- "1"
#> pre_data_object <- data_object
#> data_object <- .pre_filtering(source, data_object, "1")
#> data_object[["books"]] <- data_object[["books"]] %>%
#> dplyr::filter(author %in% c("Dan Brown", NA))
#> attr(data_object[["books"]], "filtered") <- TRUE
#> data_object[["borrowers"]] <- data_object[["borrowers"]] %>%
#> dplyr::filter((registered <= Inf & registered >= 14610) | is.na(registered))
#> attr(data_object[["borrowers"]], "filtered") <- TRUE
#> data_object <- .post_filtering(source, data_object, "1")
#> for (binding_key in binding_keys) {
#> data_object <- .run_binding(source, binding_key, pre_data_object, data_object)
#> }
#> step_id <- "2"
#> data_object <- .pre_filtering(source, data_object, "2")
#> data_object[["books"]] <- data_object[["books"]] %>%
#> dplyr::filter((copies <= 10 & copies >= 5) | is.na(copies))
#> attr(data_object[["books"]], "filtered") <- TRUE
#> data_object <- .post_filtering(source, data_object, "2")
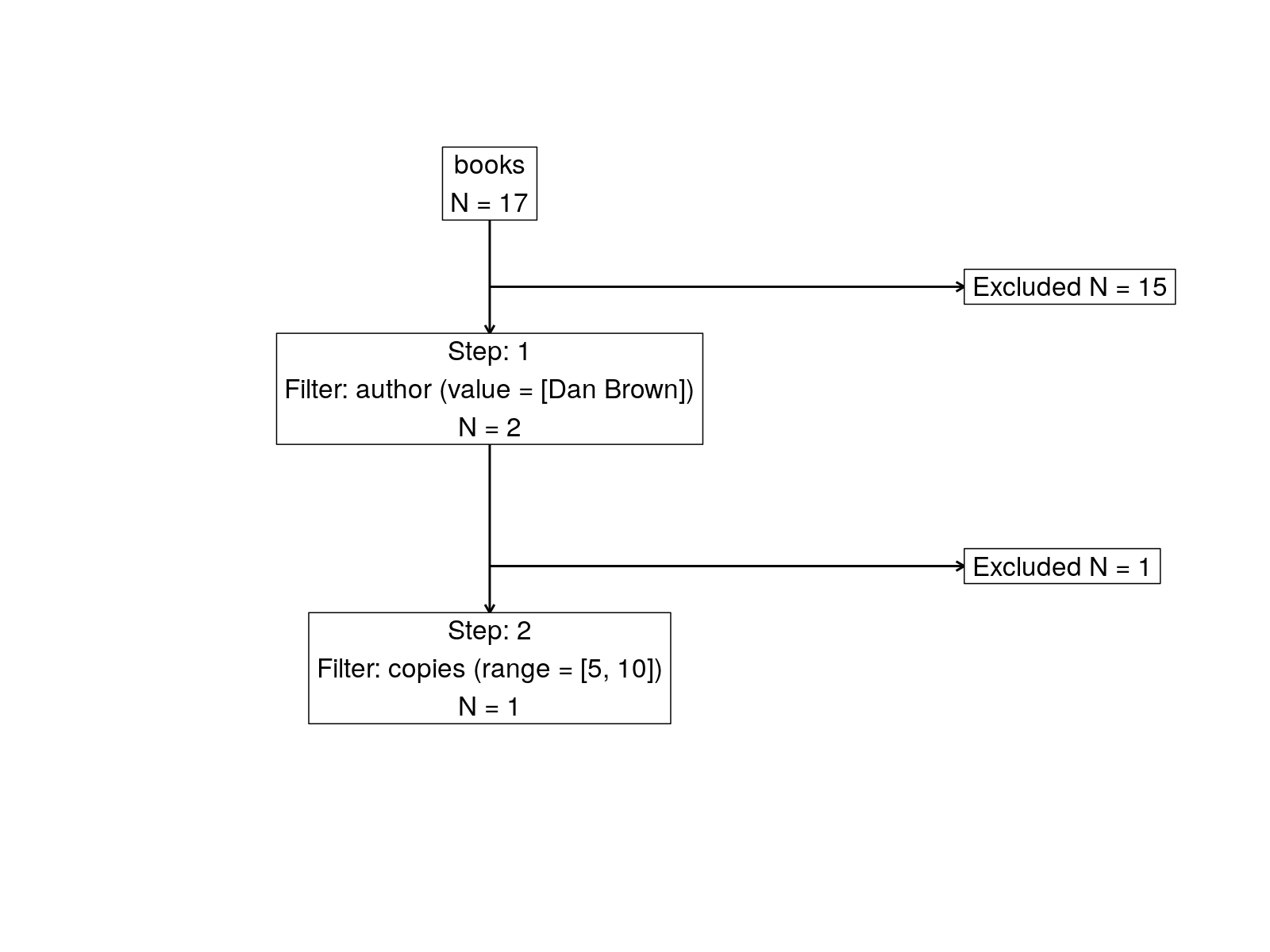
attrition(coh, dataset = "books")
get_state(coh, json = TRUE)
#> [{"step":"1","filters":[{"range":[5,6],"type":"discrete","id":"author","name":"author","variable":"author","value":"Dan Brown","dataset":"books","keep_na":true,"description":null,"active":true},{"type":"date_range","id":"registered","name":"registered","variable":"registered","range":["2010-01-01","Inf"],"dataset":"borrowers","keep_na":true,"description":null,"active":true}]},{"step":"2","filters":[{"type":"range","id":"copies","name":"copies","variable":"copies","range":[5,10],"dataset":"books","keep_na":true,"description":null,"active":true}]}]
Acknowledgement
Special thanks to:
- Kamil Wais for highlighting the need for the package and its relevance to real-world applications.
- Adam Foryś for technical support, numerous suggestions for the current and future implementation of the package.
- Paweł Kawski for indication of initial assumptions about the package based on real-world medical data.
Getting help
In a case you found any bugs, have feature request or general question please file an issue at the package Github. You may also contact the package author directly via email at [email protected].

